AFSC Eastern Bering Sea
afsc-ebs.Rmd
# Get rid of memory limits -----------------------------------------------------
options(future.globals.maxSize = 1 * 1024^4) # Allow up to 1 TB for globals
# Install Libraries ------------------------------------------------------------
# Here we list all the packages we will need for this whole process
# We'll also use this in our works cited page.
PKG <- c(
"surveyresamplr",
"dplyr",
"tidyr",
"purrr",
"ggplot2",
"future.callr",
"flextable",
"sdmTMB"
)
pkg_install <- function(p) {
if (!require(p, character.only = TRUE)) {
install.packages(p)
}
require(p, character.only = TRUE)
}
base::lapply(unique(PKG), pkg_install)
#> Loading required package: surveyresamplr
#> Loading required package: dplyr
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
#> Loading required package: tidyr
#> Loading required package: purrr
#> Loading required package: ggplot2
#> Loading required package: future.callr
#> Loading required package: future
#> Loading required package: flextable
#>
#> Attaching package: 'flextable'
#> The following object is masked from 'package:purrr':
#>
#> compose
#> Loading required package: sdmTMB
#> [[1]]
#> [1] TRUE
#>
#> [[2]]
#> [1] TRUE
#>
#> [[3]]
#> [1] TRUE
#>
#> [[4]]
#> [1] TRUE
#>
#> [[5]]
#> [1] TRUE
#>
#> [[6]]
#> [1] TRUE
#>
#> [[7]]
#> [1] TRUE
#>
#> [[8]]
#> [1] TRUE
# Set directories --------------------------------------------------------------
wd <- paste0(here::here(), "/vignettes/")
dir_out <- paste0(wd, "output/")
dir_final <- paste0(dir_out, "EBS_0results/")Run scenarios
### Define study species -------------------------------------------------------
spp_list <- data.frame(
srvy = "EBS",
common_name = c(
"walleye pollock", "snow crab", "Pacific cod",
"red king crab", "blue king crab",
"yellowfin sole", "Pacific halibut",
"Alaska plaice", "flathead sole", "northern rock sole", "arrowtooth flounder"
),
species_code = as.character(c(
21740, 68580, 21720,
69322, 69323,
10210, 10120,
10285, 10130, 10261, 10110
)),
filter_lat_lt = NA,
filter_lat_gt = NA,
filter_depth = NA,
model_fn = "total_catch_wt_kg ~ 0 + factor(year) + bottom_temperature_c",
model_family = "delta_gamma",
model_anisotropy = TRUE,
model_spatiotemporal = "iid, iid"
) |>
dplyr::mutate(
file_name = gsub(pattern = " ", replacement = "_", x = (tolower(common_name)))
)
### Load survey data -----------------------------------------------------------
# source(paste0(wd, "code/data_dl_ak.r"))
load(file = paste0(wd, "/data/noaa_afsc_catch.rda"))
catch <- noaa_afsc_catch |> dplyr::filter(srvy == "EBS")
### Load grid data -------------------------------------------------------------
load(paste0(wd, "grids/noaa_afsc_ebs_pred_grid_depth.rdata"), verbose = TRUE)
#### Add temperature: Coldpool temperature data
# Data that varies over space and time (bottom temperature)
# Here, bottom temperature, and thereby the cold pool extent, have been show to drive the distribution of many species. This is especially true for walleye pollock.
# For this we are going to lean on our in-house prepared validated and pre-prepared [{coldpool} R package](https://github.com/afsc-gap-products/coldpool) (S. Rohan, L. Barnett, and N. Charriere). This data interpolates over the whole area of the survey so there are no missing data.
crs_latlon <- "+proj=longlat +datum=WGS84" # decimal degrees
grid_yrs <-
dplyr::bind_cols(
pred_grid_depth[, c("longitude_dd", "latitude_dd", "depth_m")],
terra::unwrap(coldpool::ebs_bottom_temperature) |>
terra::project(crs_latlon) |>
terra::extract(pred_grid_depth[, c("longitude_dd", "latitude_dd")])
)
grid_yrs <- grid_yrs |>
tidyr::pivot_longer(
names_to = "year",
values_to = "bottom_temperature_c",
cols = names(grid_yrs_temperature)[4:ncol(grid_yrs_temperature)]
)
save(grid_yrs_depth_temperature, file = paste0("grids/grid_yr_temperature/noaa_afsc_ebs_pred_grid_depth_temperature.rdata"))
# # test you extracted correctkt
# ggplot(data = grid_yrs |>
# dplyr::filter(year %in% c(2022:2024)),
# mapping = ggplot2::aes(x = longitude_dd, y = latitude_dd, color = bottom_temperature_c)) +
# geom_point() +
# facet_wrap(facets = "year")
### Variables ------------------------------------------------------------------
srvy <- "EBS"
seq_from <- 0.2
seq_to <- 1.0
seq_by <- 0.2
tot_dataframes <- 13
replicate_num <- 3
### Run ------------------------------------------------------------------------
purrr::map(
1:nrow(spp_list),
~ clean_and_resample(spp_list[.x, ],
catch, seq_from, seq_to, seq_by,
tot_dataframes, replicate_num, grid_yrs, dir_out,
test = TRUE
)
)
### Plot indices ---------------------------------------------------------------
out <- plot_results(srvy = srvy, dir_out = dir_out, dir_final = dir_final)
out$plots
#> $index_boxplot_log_biomass
#>
#> $index_boxplot_log_biomass_SE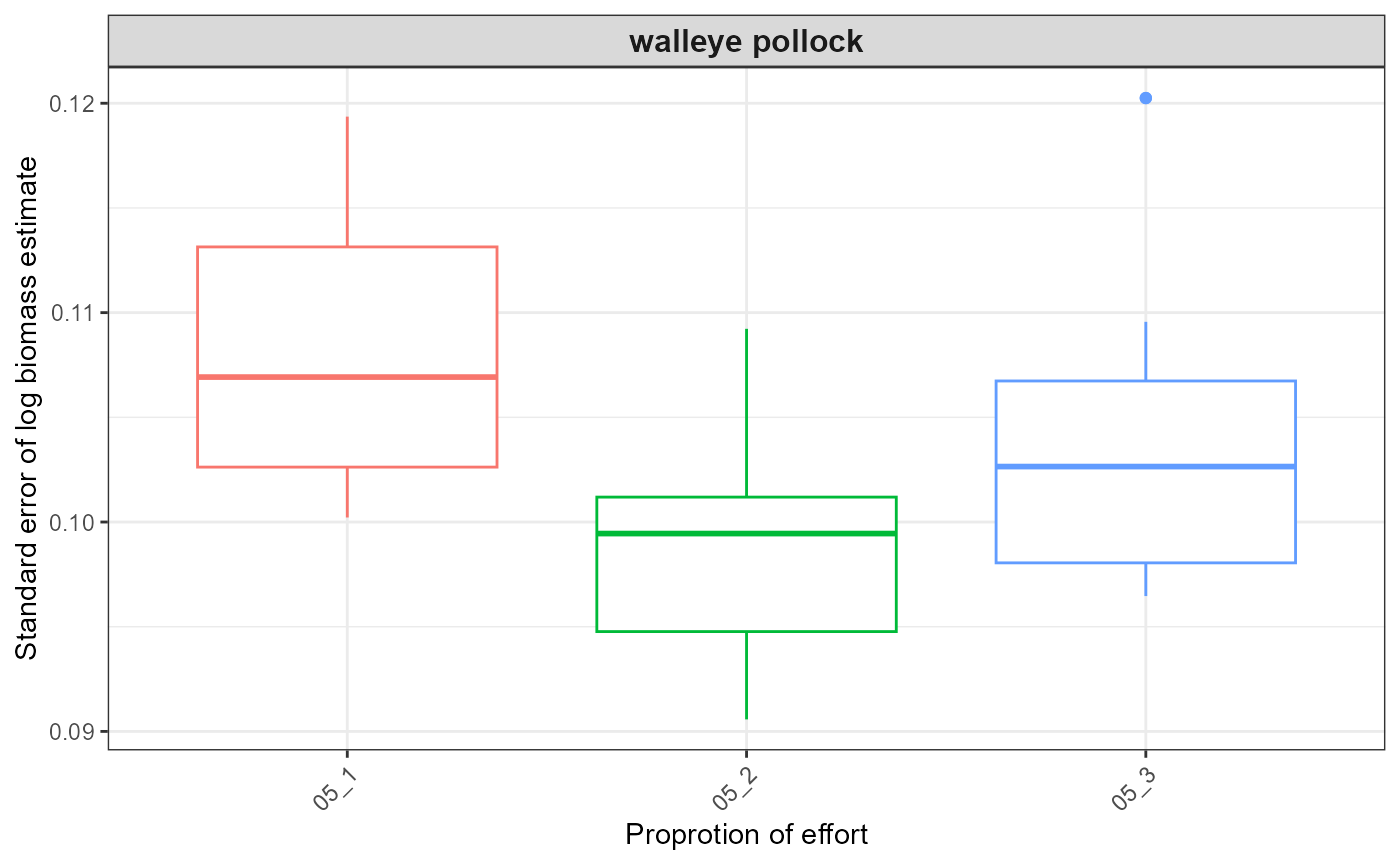
#>
#> $index_boxplot_biomass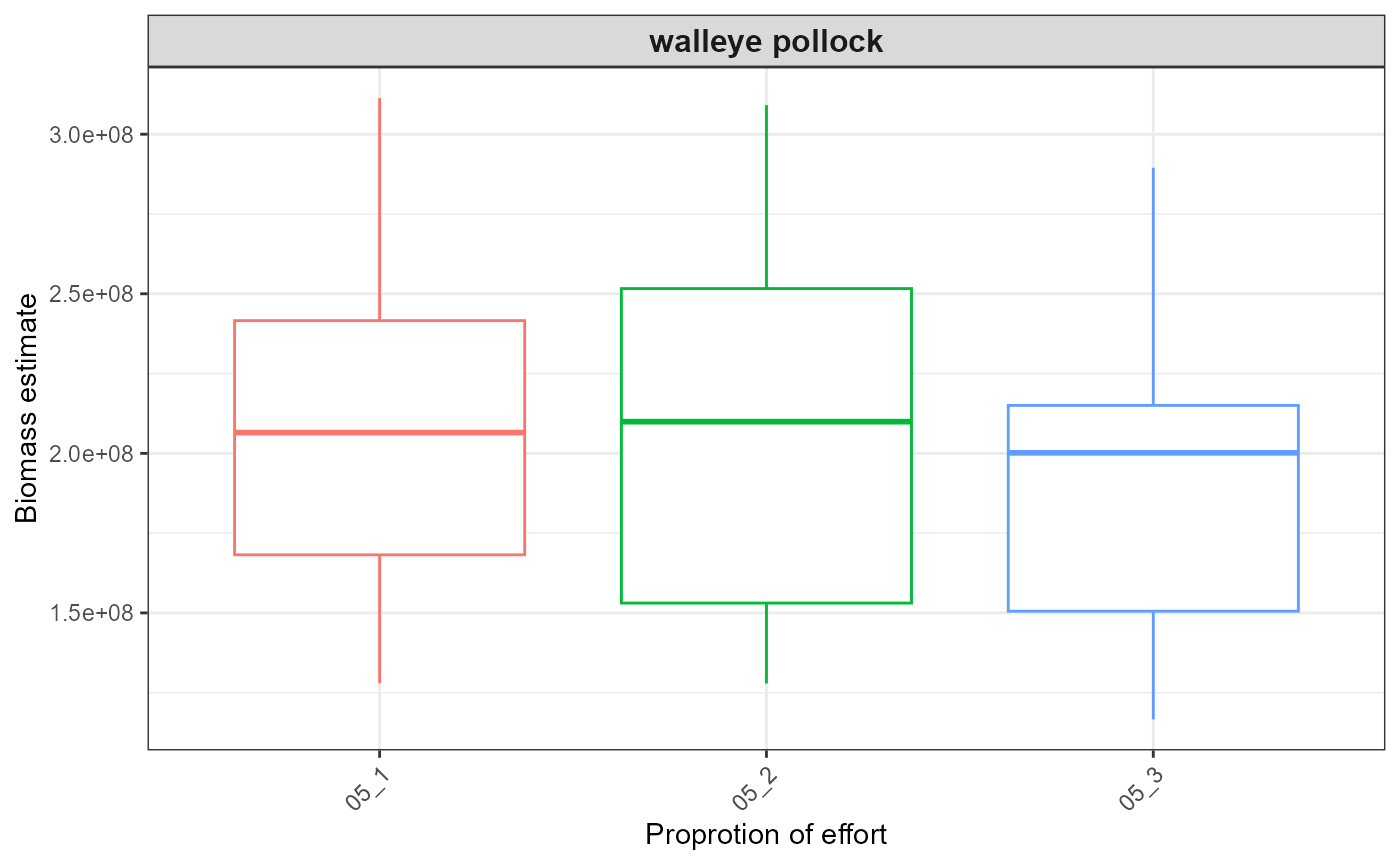
#>
#> $index_timeseries_biomass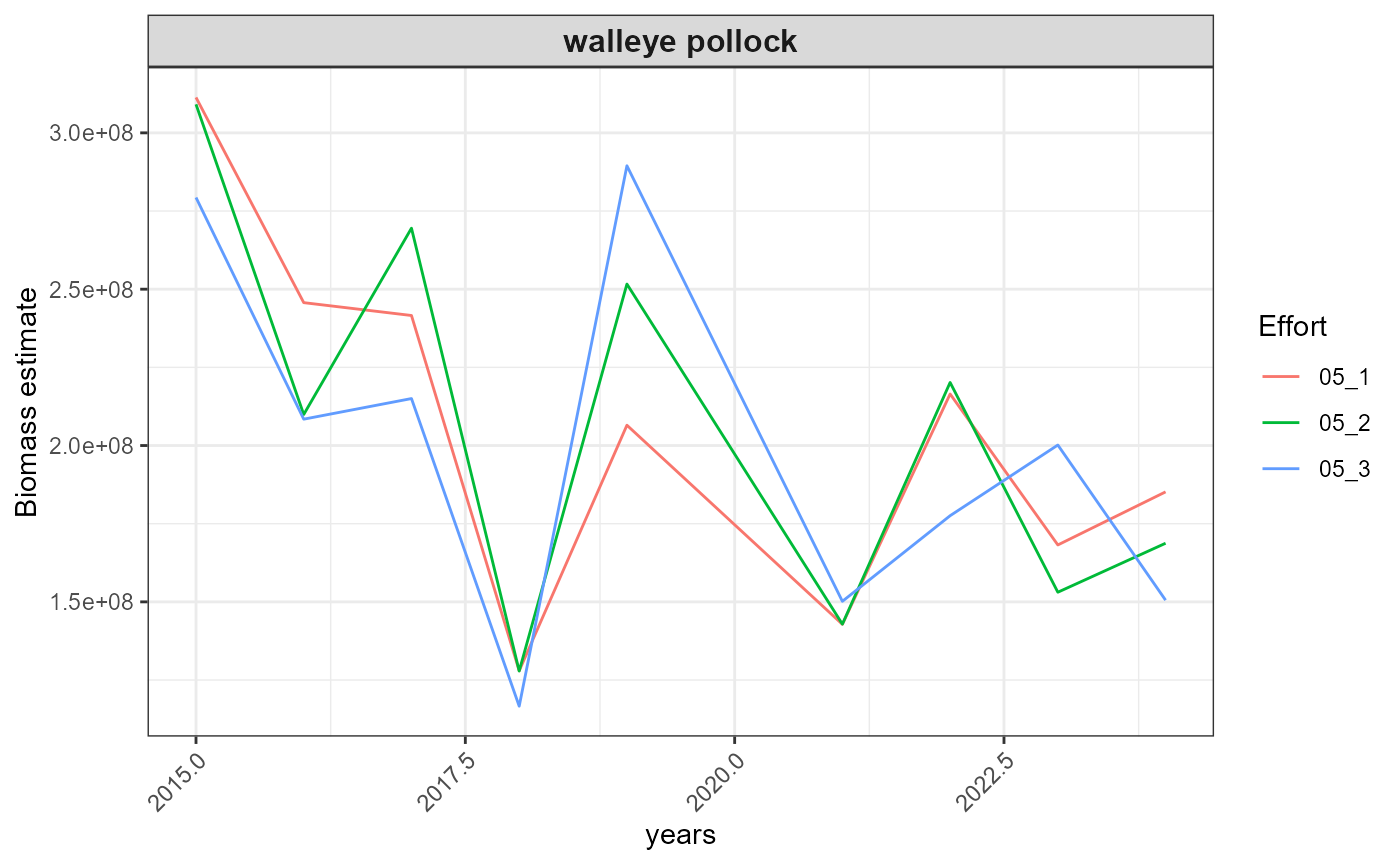
Parameter output:
i <- 1
print(names(out$tables)[i])
#> [1] "fit_df"
head(out$tables[i][[1]]) |>
flextable::flextable()X.5 |
X.4 |
X.3 |
X.2 |
X.1 |
X |
srvy |
common_name |
file_name |
species_code |
filter_lat_lt |
filter_lat_gt |
filter_depth |
model_fn |
model_family |
model_anisotropy |
model_spatiotemporal |
effort |
term |
estimate |
std.error |
conf.low |
conf.high |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
1 |
1 |
1 |
1 |
1 |
1 |
EBS |
walleye pollock |
walleye_pollock_test |
21,740 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
05_1 |
factor(year)2015 |
19.77117 |
1,391.123 |
-2,706.781 |
2,746.323 |
|||
2 |
2 |
2 |
2 |
2 |
2 |
EBS |
walleye pollock |
walleye_pollock_test |
21,740 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
05_1 |
factor(year)2016 |
19.90634 |
1,384.990 |
-2,694.625 |
2,734.437 |
|||
3 |
3 |
3 |
3 |
3 |
3 |
EBS |
walleye pollock |
walleye_pollock_test |
21,740 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
05_1 |
factor(year)2017 |
19.86007 |
1,340.321 |
-2,607.120 |
2,646.840 |
|||
4 |
4 |
4 |
4 |
4 |
4 |
EBS |
walleye pollock |
walleye_pollock_test |
21,740 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
05_1 |
factor(year)2018 |
19.80071 |
1,454.167 |
-2,830.315 |
2,869.916 |
|||
5 |
5 |
5 |
5 |
5 |
5 |
EBS |
walleye pollock |
walleye_pollock_test |
21,740 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
05_1 |
factor(year)2019 |
20.11844 |
1,272.466 |
-2,473.869 |
2,514.106 |
|||
6 |
6 |
6 |
6 |
6 |
6 |
EBS |
walleye pollock |
walleye_pollock_test |
21,740 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
05_1 |
factor(year)2021 |
19.71943 |
1,278.253 |
-2,485.611 |
2,525.050 |
i <- 1 + i
print(names(out$tables)[i])
#> [1] "fit_pars"
head(out$tables[i][[1]]) |>
flextable::flextable()X.5 |
X.4 |
X.3 |
X.2 |
X.1 |
X |
srvy |
common_name |
file_name |
species_code |
filter_lat_lt |
filter_lat_gt |
filter_depth |
model_fn |
model_family |
model_anisotropy |
model_spatiotemporal |
effort |
term |
estimate |
std.error |
conf.low |
conf.high |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
1 |
1 |
1 |
1 |
1 |
1 |
EBS |
walleye pollock |
walleye_pollock_test |
21,740 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
05_1 |
range |
3.7298099 |
3,393.6903 |
0 |
Inf |
|||
2 |
2 |
2 |
2 |
2 |
2 |
EBS |
walleye pollock |
walleye_pollock_test |
21,740 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
05_1 |
sigma_O |
0.1812892 |
140.5073 |
0 |
Inf |
|||
3 |
3 |
3 |
3 |
3 |
3 |
EBS |
walleye pollock |
walleye_pollock_test |
21,740 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
05_1 |
sigma_E |
0.3717503 |
336.2267 |
0 |
Inf |
|||
4 |
4 |
4 |
4 |
4 |
EBS |
walleye pollock |
walleye_pollock_test |
21,740 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
05_2 |
range |
2.8284007 |
|||||||
5 |
5 |
5 |
5 |
5 |
EBS |
walleye pollock |
walleye_pollock_test |
21,740 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
05_2 |
sigma_O |
0.2820909 |
|||||||
6 |
6 |
6 |
6 |
6 |
EBS |
walleye pollock |
walleye_pollock_test |
21,740 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
05_2 |
sigma_E |
0.2820909 |
i <- 1 + i
print(names(out$tables)[i])
#> [1] "fit_check"
head(out$tables[i][[1]]) |>
flextable::flextable()X.5 |
X.4 |
X.3 |
X.2 |
X.1 |
X |
srvy |
common_name |
file_name |
species_code |
filter_lat_lt |
filter_lat_gt |
filter_depth |
model_fn |
model_family |
model_anisotropy |
model_spatiotemporal |
effort |
hessian_ok |
eigen_values_ok |
nlminb_ok |
range_ok |
gradients_ok |
se_magnitude_ok |
se_na_ok |
sigmas_ok |
all_ok |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
1 |
1 |
1 |
1 |
1 |
1 |
EBS |
walleye pollock |
walleye_pollock_test |
21,740 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
05_1 |
FALSE |
TRUE |
TRUE |
TRUE |
TRUE |
FALSE |
TRUE |
TRUE |
FALSE |
|||
2 |
2 |
2 |
2 |
2 |
EBS |
walleye pollock |
walleye_pollock_test |
21,740 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
05_2 |
FALSE |
TRUE |
TRUE |
TRUE |
FALSE |
FALSE |
TRUE |
TRUE |
FALSE |
||||
3 |
3 |
3 |
3 |
EBS |
walleye pollock |
walleye_pollock_test |
21,740 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
05_3 |
FALSE |
TRUE |
TRUE |
TRUE |
FALSE |
FALSE |
TRUE |
TRUE |
FALSE |
|||||
4 |
4 |
4 |
EBS |
walleye pollock |
simple_walleye_pollock |
21,740 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
05_1 |
FALSE |
TRUE |
TRUE |
TRUE |
TRUE |
FALSE |
TRUE |
TRUE |
FALSE |
||||||
5 |
5 |
EBS |
walleye pollock |
simple_walleye_pollock |
21,740 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
05_2 |
FALSE |
TRUE |
TRUE |
TRUE |
FALSE |
FALSE |
TRUE |
TRUE |
FALSE |
|||||||
6 |
EBS |
walleye pollock |
simple_walleye_pollock |
21,740 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
05_3 |
FALSE |
TRUE |
TRUE |
TRUE |
FALSE |
FALSE |
TRUE |
TRUE |
FALSE |
i <- 1 + i
print(names(out$tables)[i])
#> [1] "index"
head(out$tables[i][[1]]) |>
flextable::flextable()X.5 |
X.4 |
X.3 |
X.2 |
X.1 |
X |
srvy |
common_name |
file_name |
species_code |
filter_lat_lt |
filter_lat_gt |
filter_depth |
model_fn |
model_family |
model_anisotropy |
model_spatiotemporal |
effort |
year |
est |
lwr |
upr |
log_est |
se |
type |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
1 |
1 |
1 |
1 |
1 |
1 |
EBS |
walleye pollock |
walleye_pollock_test |
21,740 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
05_1 |
2,015 |
311,329,658 |
254,290,109 |
381,163,688 |
19.55636 |
0.1032552 |
index |
|||
2 |
2 |
2 |
2 |
2 |
2 |
EBS |
walleye pollock |
walleye_pollock_test |
21,740 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
05_1 |
2,016 |
245,697,312 |
201,795,592 |
299,150,087 |
19.31961 |
0.1004330 |
index |
|||
3 |
3 |
3 |
3 |
3 |
3 |
EBS |
walleye pollock |
walleye_pollock_test |
21,740 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
05_1 |
2,017 |
241,587,105 |
198,504,083 |
294,020,799 |
19.30274 |
0.1002163 |
index |
|||
4 |
4 |
4 |
4 |
4 |
4 |
EBS |
walleye pollock |
walleye_pollock_test |
21,740 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
05_1 |
2,018 |
127,954,658 |
101,264,147 |
161,680,073 |
18.66719 |
0.1193611 |
index |
|||
5 |
5 |
5 |
5 |
5 |
5 |
EBS |
walleye pollock |
walleye_pollock_test |
21,740 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
05_1 |
2,019 |
206,496,853 |
166,663,257 |
255,850,936 |
19.14580 |
0.1093438 |
index |
|||
6 |
6 |
6 |
6 |
6 |
6 |
EBS |
walleye pollock |
walleye_pollock_test |
21,740 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
05_1 |
2,021 |
142,822,421 |
115,820,004 |
176,120,214 |
18.77711 |
0.1069228 |
index |