AFSC Aleutian Islands
afsc-ai.Rmd
# Get rid of memory limits -----------------------------------------------------
options(future.globals.maxSize = 1 * 1024^4) # Allow up to 1 TB for globals
# Install Libraries ------------------------------------------------------------
# Here we list all the packages we will need for this whole process
# We'll also use this in our works cited page.
PKG <- c(
"surveyresamplr",
"dplyr",
"tidyr",
"purrr",
"ggplot2",
"future.callr",
"flextable",
"sdmTMB"
)
pkg_install <- function(p) {
if (!require(p, character.only = TRUE)) {
install.packages(p)
}
require(p, character.only = TRUE)
}
base::lapply(unique(PKG), pkg_install)
#> Loading required package: surveyresamplr
#> Loading required package: dplyr
#>
#> Attaching package: 'dplyr'
#> The following objects are masked from 'package:stats':
#>
#> filter, lag
#> The following objects are masked from 'package:base':
#>
#> intersect, setdiff, setequal, union
#> Loading required package: tidyr
#> Loading required package: purrr
#> Loading required package: ggplot2
#> Loading required package: future.callr
#> Loading required package: future
#> Loading required package: flextable
#>
#> Attaching package: 'flextable'
#> The following object is masked from 'package:purrr':
#>
#> compose
#> Loading required package: sdmTMB
#> [[1]]
#> [1] TRUE
#>
#> [[2]]
#> [1] TRUE
#>
#> [[3]]
#> [1] TRUE
#>
#> [[4]]
#> [1] TRUE
#>
#> [[5]]
#> [1] TRUE
#>
#> [[6]]
#> [1] TRUE
#>
#> [[7]]
#> [1] TRUE
#>
#> [[8]]
#> [1] TRUE
# Set directories --------------------------------------------------------------
wd <- paste0(here::here(), "/vignettes/")
dir_out <- paste0(wd, "output/")
dir_final <- paste0(dir_out, "AI_0results/")Run scenarios
### Define study species -------------------------------------------------------
spp_list <- data.frame(
srvy = "AI",
common_name = c(
"walleye pollock", "Pacific cod",
"Pacific ocean perch", "flathead sole",
"northern rockfish", "arrowtooth flounder"
),
species_code = as.character(c(
21740, 21720,
30060, 10130,
30420, 10110
)),
filter_lat_lt = NA,
filter_lat_gt = NA,
filter_depth = NA,
model_fn = "total_catch_wt_kg ~ 0 + factor(year)",
model_family = "delta_gamma",
model_anisotropy = TRUE,
model_spatiotemporal = "iid, iid"
) |>
dplyr::mutate(
file_name = gsub(pattern = " ", replacement = "_", x = (tolower(common_name)))
)
### Load survey data -----------------------------------------------------------
# source(paste0(wd, "code/data_dl_ak.r"))
catch <- surveyresamplr::noaa_afsc_catch |>
dplyr::filter(srvy == "AI")
### Load grid data -------------------------------------------------------------
grid_yrs <- sdmTMB::replicate_df(surveyresamplr::noaa_afsc_ai_pred_grid_depth, "year", unique(catch$year))
### Variables ------------------------------------------------------------------
srvy <- "AI"
seq_from <- 0.2
seq_to <- 1.0
seq_by <- 0.2
tot_dataframes <- 13
replicate_num <- 3
### Run ------------------------------------------------------------------------
map(
1:nrow(spp_list),
~ clean_and_resample(spp_list[.x, ],
catch, seq_from, seq_to, seq_by,
tot_dataframes, replicate_num, grid_yrs, dir_out,
test = FALSE
)
)
### Plot indices --------------------------------------------------------------
out <- plot_results(srvy = srvy, dir_out = dir_out, dir_final = dir_final)
out$plots
#> $index_boxplot_log_biomass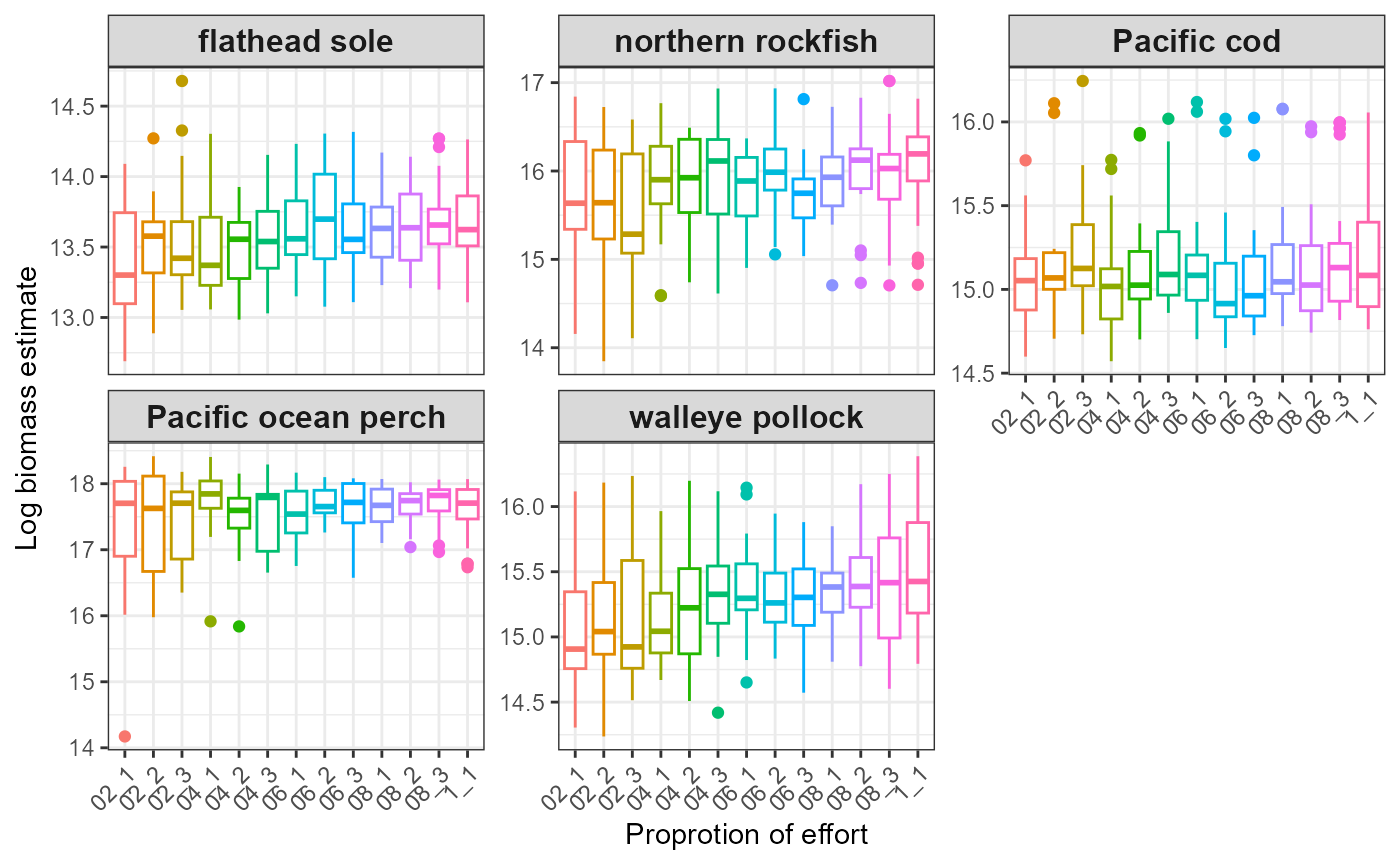
#>
#> $index_boxplot_log_biomass_SE
#> Warning: Removed 14 rows containing non-finite outside the scale range
#> (`stat_boxplot()`).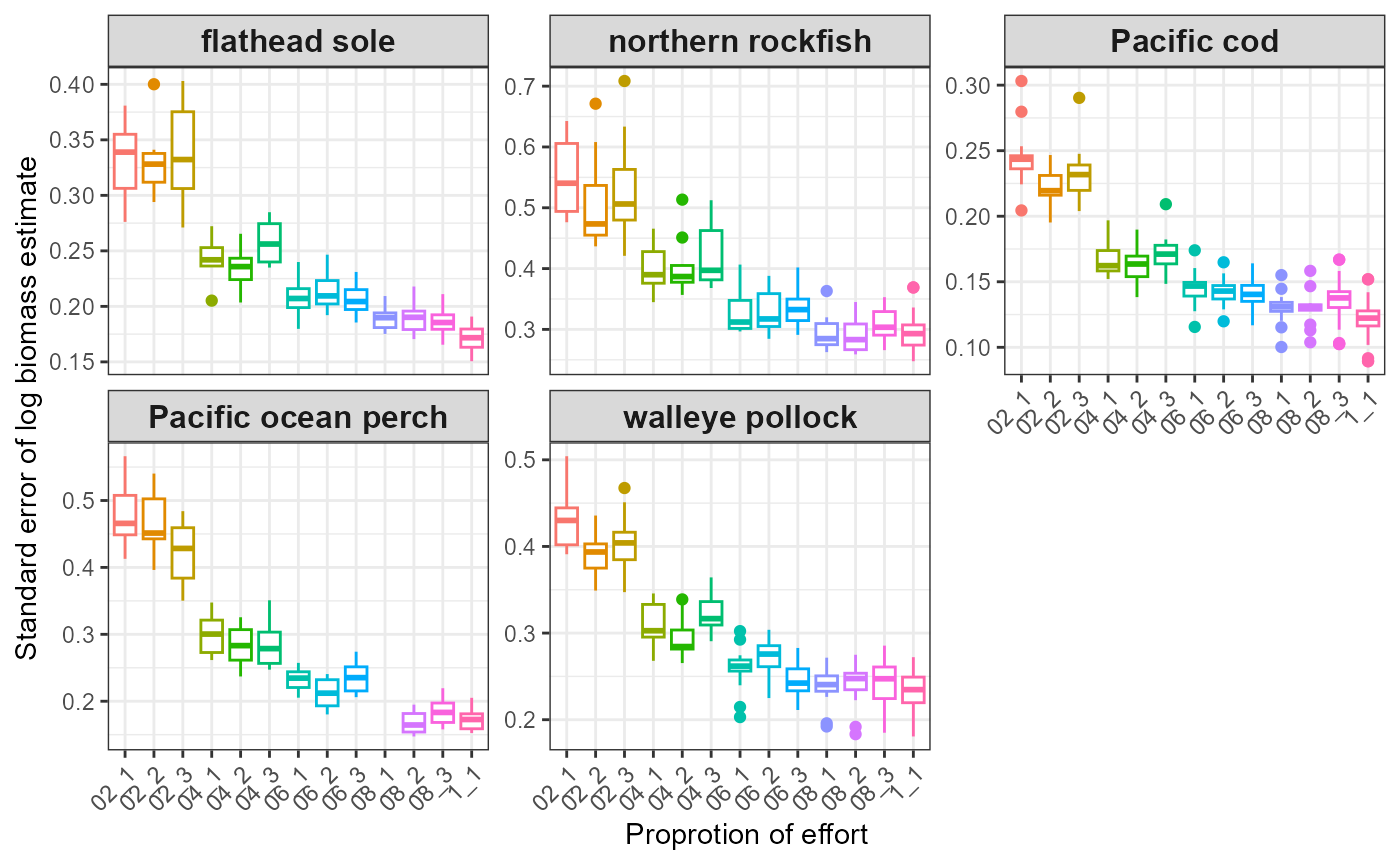
#>
#> $index_boxplot_biomass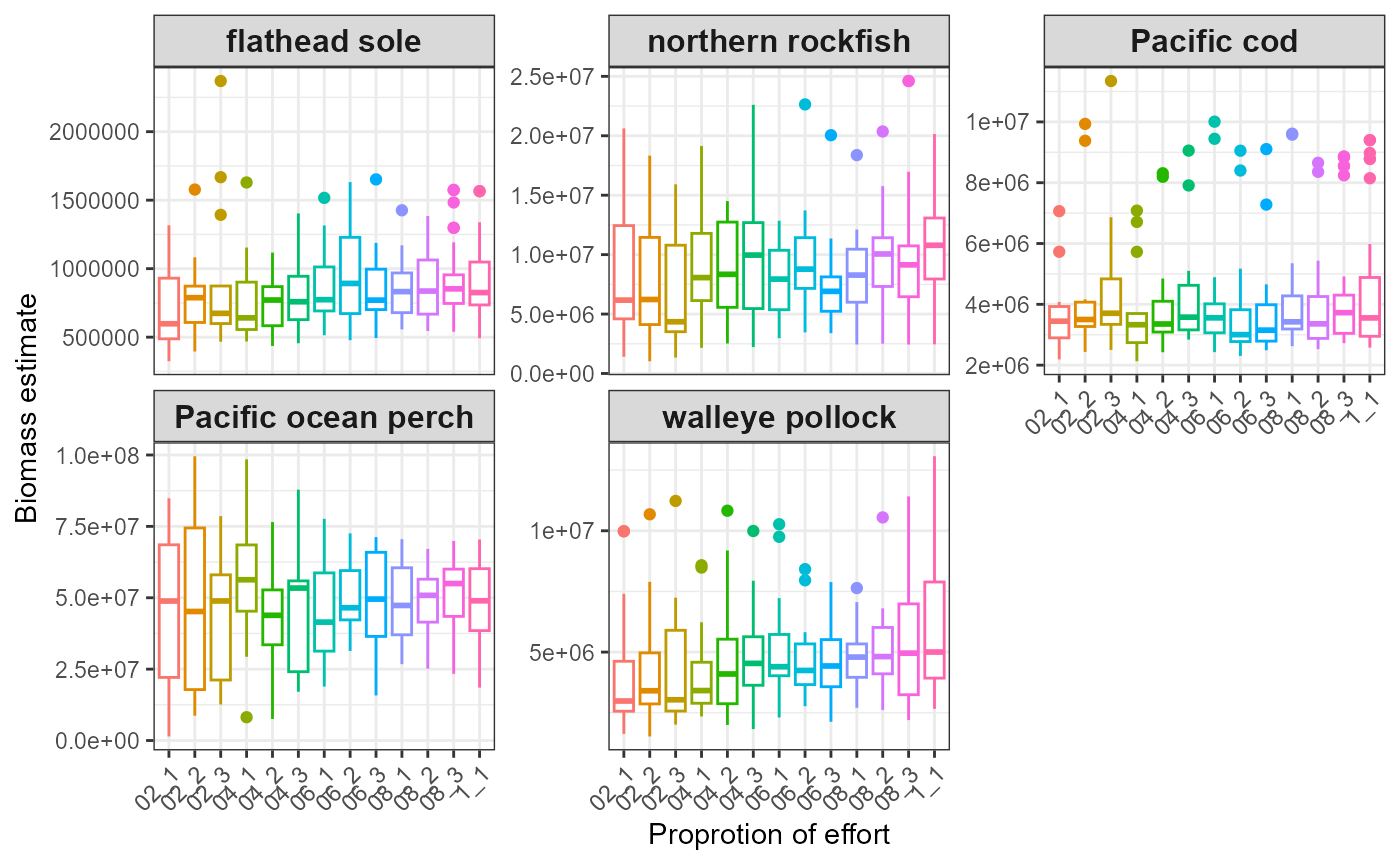
#>
#> $index_timeseries_biomass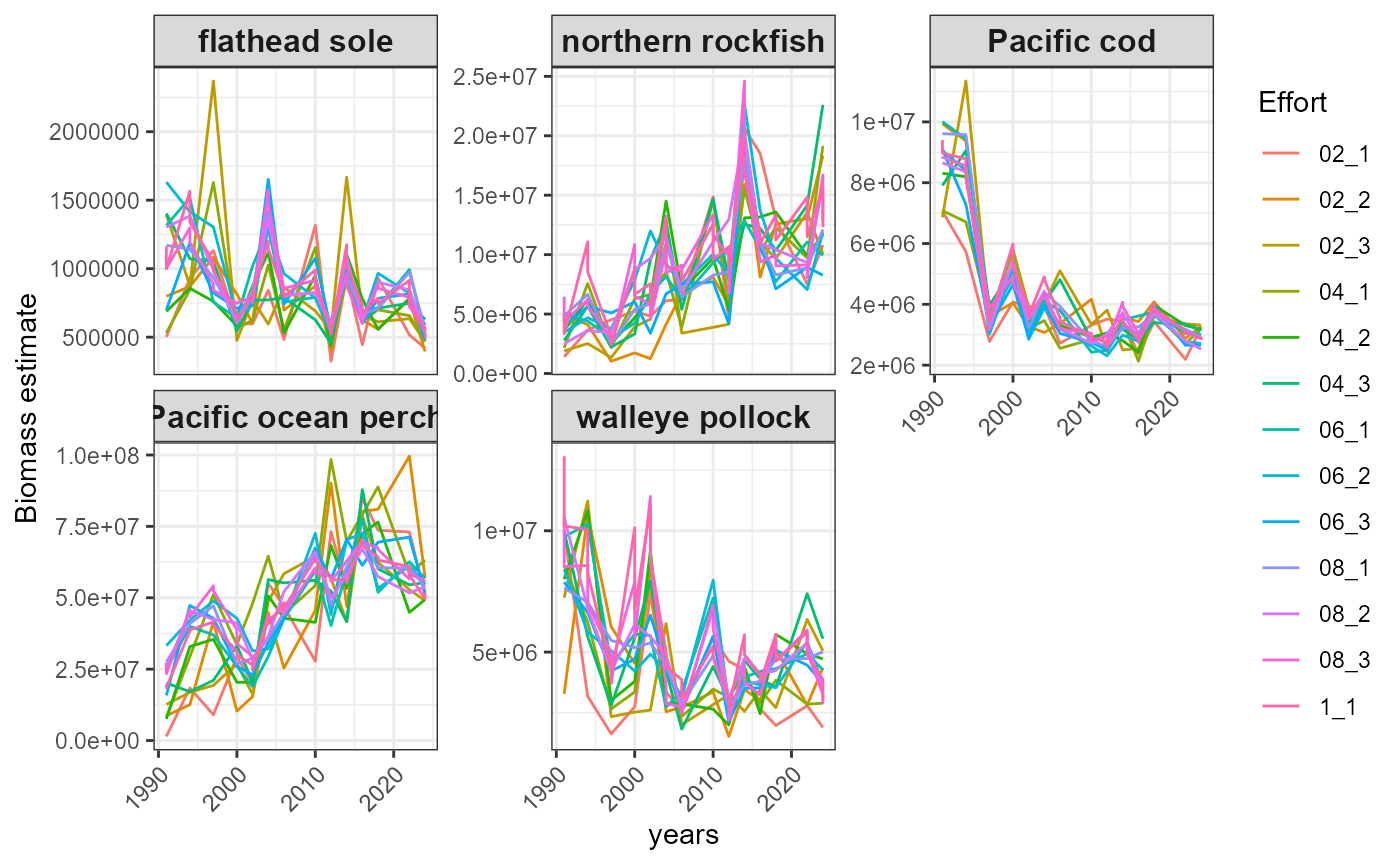
Parameter output:
i <- 1
print(names(out$tables)[i])
#> [1] "fit_df"
head(out$tables[i][[1]]) |>
flextable::flextable()X.16 |
X.15 |
X.14 |
X.13 |
X.12 |
X.11 |
X.10 |
X.9 |
X.8 |
X.7 |
X.6 |
X.5 |
X.4 |
X.3 |
X.2 |
X.1 |
X |
srvy |
common_name |
species_code |
filter_lat_lt |
filter_lat_gt |
filter_depth |
model_fn |
model_family |
model_anisotropy |
model_spatiotemporal |
file_name |
effort |
term |
estimate |
std.error |
conf.low |
conf.high |
X.17 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
AI |
flathead sole |
10,130 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
flathead_sole |
08_3 |
factor(year)1991 |
18.86459 |
1,396.8455 |
-2,718.902 |
2,756.631 |
||||
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
AI |
flathead sole |
10,130 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
flathead_sole |
08_3 |
factor(year)1994 |
18.61412 |
1,055.3335 |
-2,049.802 |
2,087.030 |
||||
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
AI |
flathead sole |
10,130 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
flathead_sole |
08_3 |
factor(year)1997 |
18.35688 |
955.0542 |
-1,853.515 |
1,890.229 |
||||
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
AI |
flathead sole |
10,130 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
flathead_sole |
08_3 |
factor(year)2000 |
19.33032 |
1,468.8298 |
-2,859.523 |
2,898.184 |
||||
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
AI |
flathead sole |
10,130 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
flathead_sole |
08_3 |
factor(year)2002 |
20.10074 |
2,321.0138 |
-4,529.003 |
4,569.204 |
||||
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
AI |
flathead sole |
10,130 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
flathead_sole |
08_3 |
factor(year)2004 |
20.22360 |
2,279.3796 |
-4,447.278 |
4,487.726 |
i <- 1 + i
print(names(out$tables)[i])
#> [1] "fit_pars"
head(out$tables[i][[1]]) |>
flextable::flextable()X.16 |
X.15 |
X.14 |
X.13 |
X.12 |
X.11 |
X.10 |
X.9 |
X.8 |
X.7 |
X.6 |
X.5 |
X.4 |
X.3 |
X.2 |
X.1 |
X |
srvy |
common_name |
species_code |
filter_lat_lt |
filter_lat_gt |
filter_depth |
model_fn |
model_family |
model_anisotropy |
model_spatiotemporal |
file_name |
effort |
term |
estimate |
std.error |
conf.low |
conf.high |
X.17 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
AI |
flathead sole |
10,130 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
flathead_sole |
08_3 |
range |
0.5012001 |
|||||||
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
AI |
flathead sole |
10,130 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
flathead_sole |
08_3 |
sigma_O |
0.4811169 |
|||||||
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
AI |
flathead sole |
10,130 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
flathead_sole |
08_3 |
sigma_E |
0.2237463 |
187.6216 |
0 |
Inf |
||||
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
AI |
flathead sole |
10,130 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
flathead_sole |
1_1 |
range |
2.8260651 |
||||||||
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
AI |
flathead sole |
10,130 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
flathead_sole |
1_1 |
sigma_O |
0.2817437 |
||||||||
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
AI |
flathead sole |
10,130 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
flathead_sole |
1_1 |
sigma_E |
0.2817441 |
i <- 1 + i
print(names(out$tables)[i])
#> [1] "fit_check"
head(out$tables[i][[1]]) |>
flextable::flextable()X.16 |
X.15 |
X.14 |
X.13 |
X.12 |
X.11 |
X.10 |
X.9 |
X.8 |
X.7 |
X.6 |
X.5 |
X.4 |
X.3 |
X.2 |
X.1 |
X |
srvy |
common_name |
species_code |
filter_lat_lt |
filter_lat_gt |
filter_depth |
model_fn |
model_family |
model_anisotropy |
model_spatiotemporal |
file_name |
effort |
hessian_ok |
eigen_values_ok |
nlminb_ok |
range_ok |
gradients_ok |
se_magnitude_ok |
se_na_ok |
sigmas_ok |
all_ok |
X.17 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
AI |
flathead sole |
10,130 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
flathead_sole |
08_3 |
FALSE |
TRUE |
TRUE |
TRUE |
TRUE |
FALSE |
TRUE |
FALSE |
FALSE |
||||
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
AI |
flathead sole |
10,130 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
flathead_sole |
1_1 |
FALSE |
TRUE |
TRUE |
TRUE |
FALSE |
FALSE |
TRUE |
FALSE |
FALSE |
|||||
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
AI |
flathead sole |
10,130 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
flathead_sole |
08_3 |
FALSE |
TRUE |
TRUE |
TRUE |
TRUE |
FALSE |
TRUE |
FALSE |
FALSE |
||||||
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
AI |
flathead sole |
10,130 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
flathead_sole |
1_1 |
FALSE |
TRUE |
TRUE |
TRUE |
FALSE |
FALSE |
TRUE |
FALSE |
FALSE |
|||||||
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
AI |
flathead sole |
10,130 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
flathead_sole |
02_1 |
FALSE |
TRUE |
TRUE |
TRUE |
TRUE |
FALSE |
TRUE |
FALSE |
FALSE |
||||||||
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
AI |
flathead sole |
10,130 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
flathead_sole |
02_2 |
FALSE |
TRUE |
TRUE |
TRUE |
TRUE |
FALSE |
TRUE |
FALSE |
FALSE |
i <- 1 + i
print(names(out$tables)[i])
#> [1] "index"
head(out$tables[i][[1]]) |>
flextable::flextable()X.16 |
X.15 |
X.14 |
X.13 |
X.12 |
X.11 |
X.10 |
X.9 |
X.8 |
X.7 |
X.6 |
X.5 |
X.4 |
X.3 |
X.2 |
X.1 |
X |
srvy |
common_name |
species_code |
filter_lat_lt |
filter_lat_gt |
filter_depth |
model_fn |
model_family |
model_anisotropy |
model_spatiotemporal |
file_name |
effort |
year |
est |
lwr |
upr |
log_est |
se |
type |
X.17 |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
1 |
AI |
flathead sole |
10,130 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
flathead_sole |
08_3 |
1,991 |
1,003,367.7 |
675,616.5 |
1,490,116 |
13.81887 |
0.2017852 |
index |
||||
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
2 |
AI |
flathead sole |
10,130 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
flathead_sole |
08_3 |
1,994 |
1,298,209.7 |
894,878.5 |
1,883,327 |
14.07650 |
0.1898267 |
index |
||||
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
3 |
AI |
flathead sole |
10,130 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
flathead_sole |
08_3 |
1,997 |
887,367.2 |
608,595.0 |
1,293,834 |
13.69601 |
0.1924045 |
index |
||||
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
4 |
AI |
flathead sole |
10,130 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
flathead_sole |
08_3 |
2,000 |
746,057.9 |
520,022.3 |
1,070,343 |
13.52256 |
0.1841521 |
index |
||||
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
5 |
AI |
flathead sole |
10,130 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
flathead_sole |
08_3 |
2,002 |
792,123.4 |
534,154.3 |
1,174,678 |
13.58247 |
0.2010406 |
index |
||||
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
6 |
AI |
flathead sole |
10,130 |
total_catch_wt_kg ~ 0 + factor(year) |
delta_gamma |
TRUE |
iid, iid |
flathead_sole |
08_3 |
2,004 |
1,574,598.9 |
1,095,567.7 |
2,263,084 |
14.26951 |
0.1850687 |
index |