OSA Tutorial
Andrea Havron
2025-06-25
TMB-validation-tutorial.RmdMethods
Examples will reply on two models from the TMB example suite:
- simple.cpp: normal linear mixed model
- arxar1.cpp: generalized linear mixed model with separable covariance on lattice with AR1 structure in each direction
Examples can be run in R using:
TMB::runExample(name = "simple") and
TMB::runExample(name = "ar1xar1")
Assumption #1: Data and random effects are approximately normal
path <- here::here()
TMB::runExample(name = "randomwalkvalidation", exfolder = paste0(path,"/inst/examples"))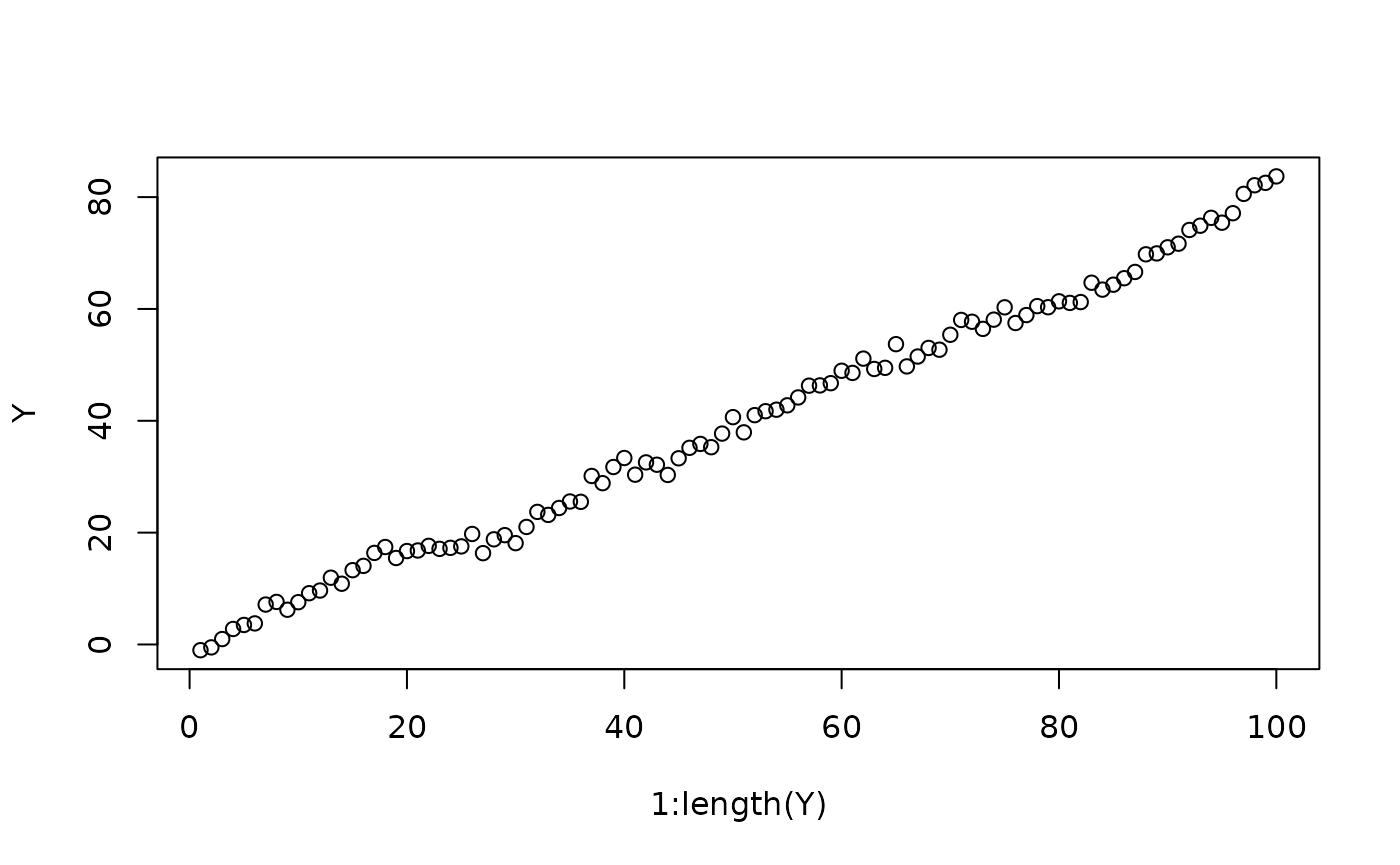
Full Guassian
#Correct Model
osa <- TMB::oneStepPredict(obj1, observation.name = "y", method = "fullGaussian")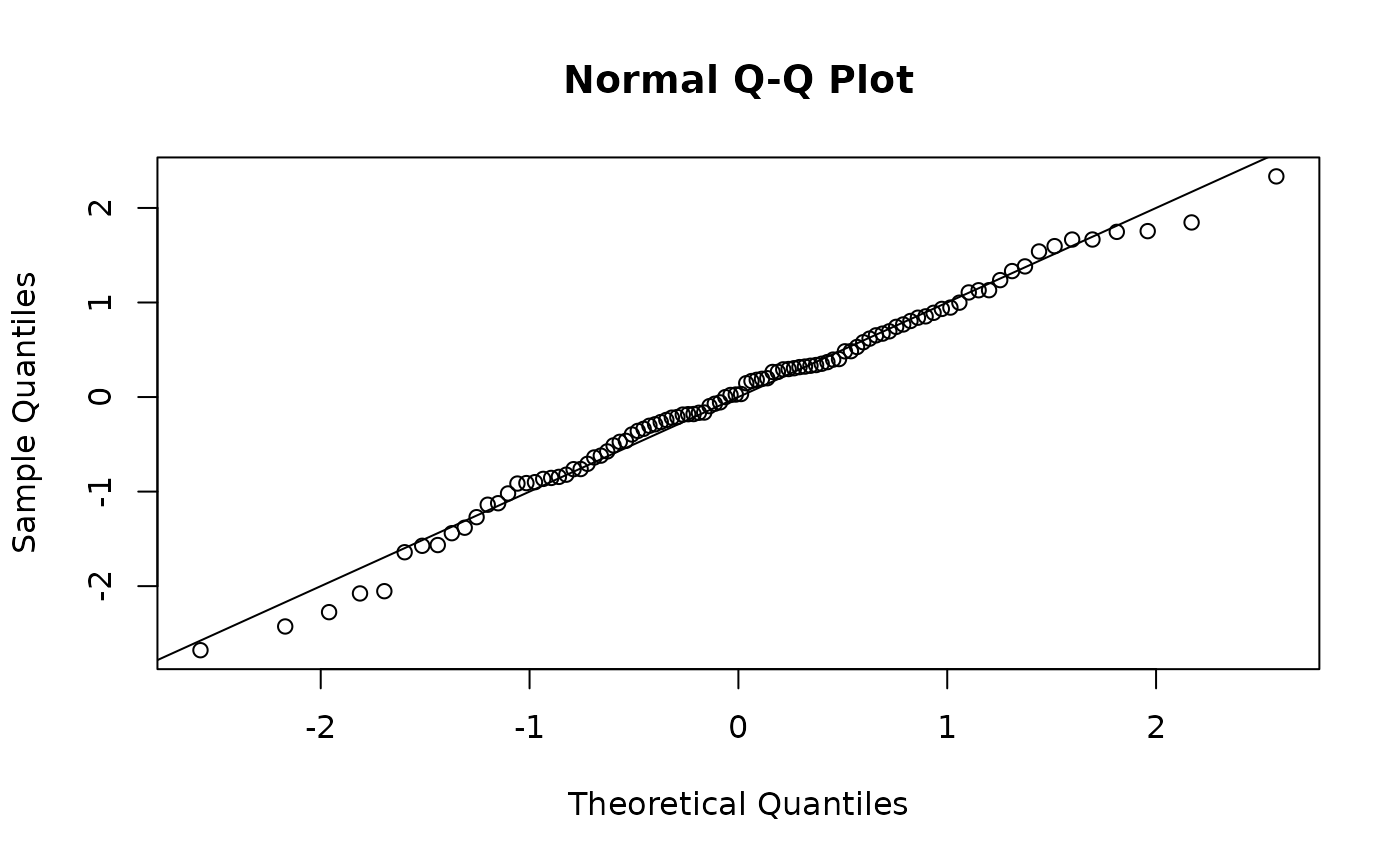
#Mis-specified Model
osa <- TMB::oneStepPredict(obj0, observation.name = "y", method = "fullGaussian")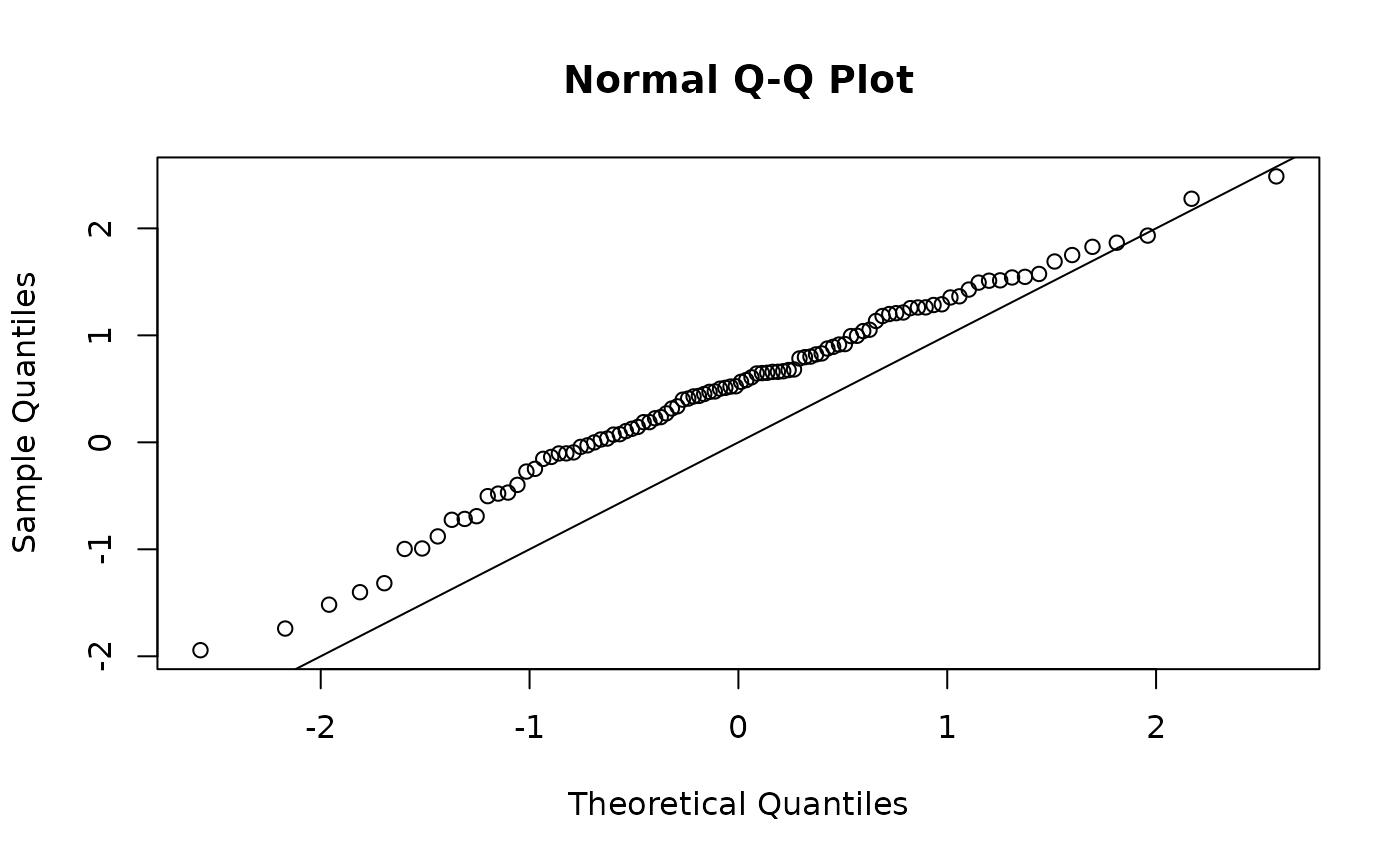
one-step Guassian
osa <- TMB::oneStepPredict(obj1, observation.name = "y", data.term.indicator = "keep",
method = "oneStepGaussian")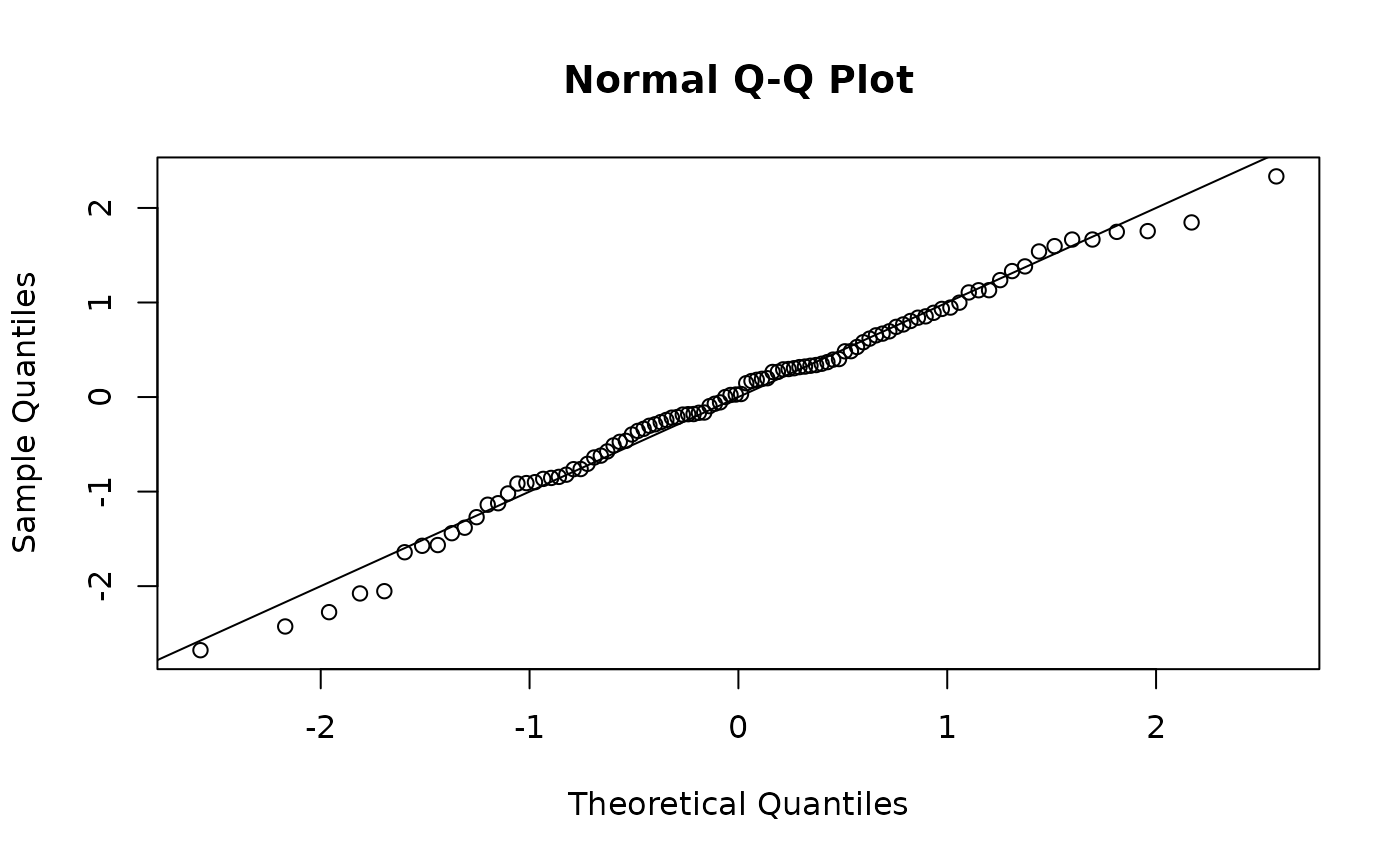
Assumption #2: Data are not approximately normal but have a defined cdf
#Plot data:
ggplot2::ggplot(d, ggplot2::aes(x,y,fill = N)) + ggplot2::geom_raster()
Data are discrete, so need to set discrete = TRUE

