Results - KS Test
TMB-validation-figures-results-phylo.RmdPhylogenetic Covariance Structure
Linear Mixed Model
Theoretical Residuals
Kolmogorov-Smirnov Test
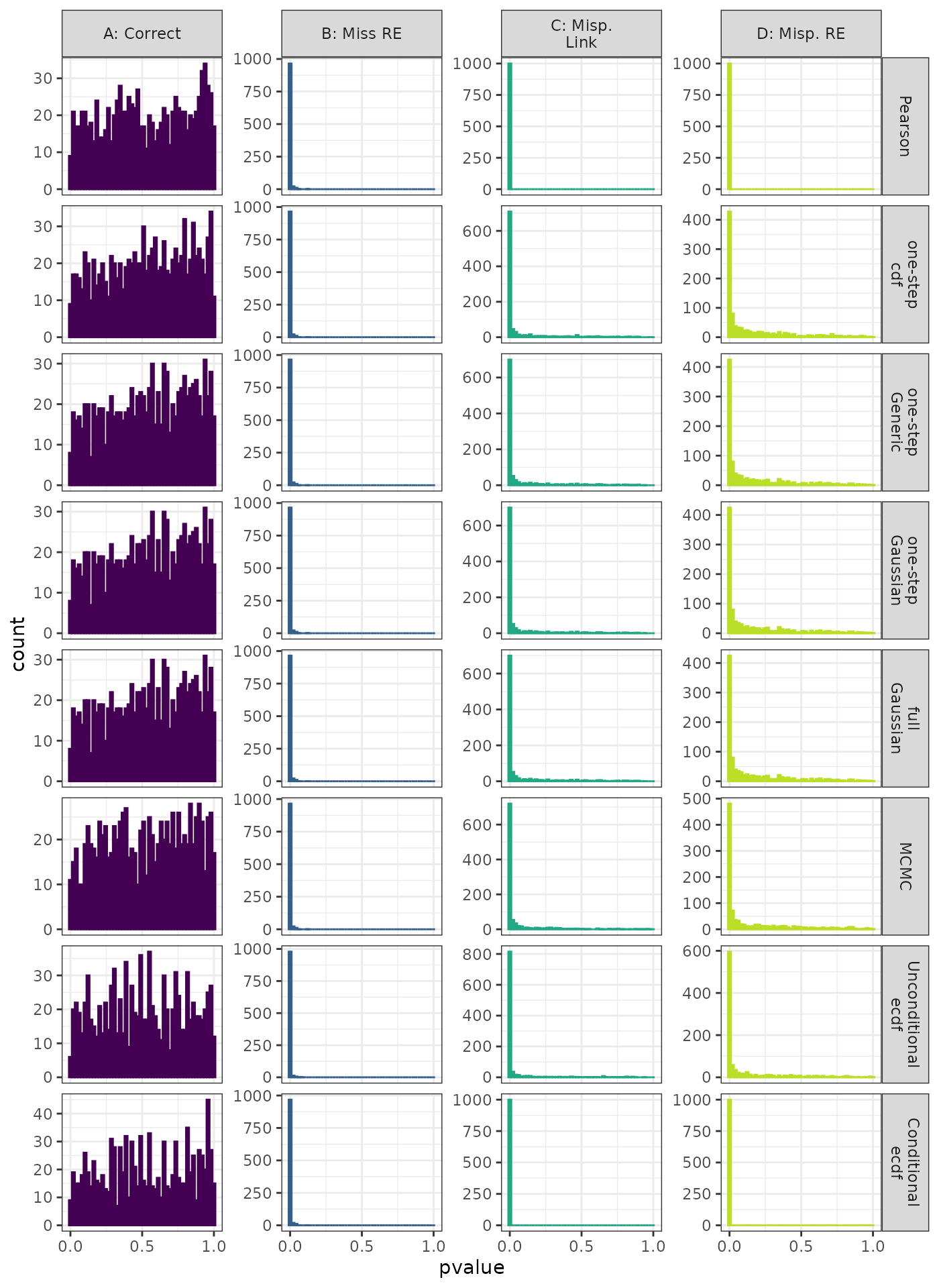
Linear Mixed Model. Distribution of theoretical p-values under the correct (A) and mis-specified (B-D) models evaluated under the Kolmogorov-Smirnov Test for each method when true parameters are known. Mis-specification are, from left to right: Missing Random Effect (B), Log-linked data fit to Normal (C), Mis-specified Random Effect (D).
Anderson-Darling Test
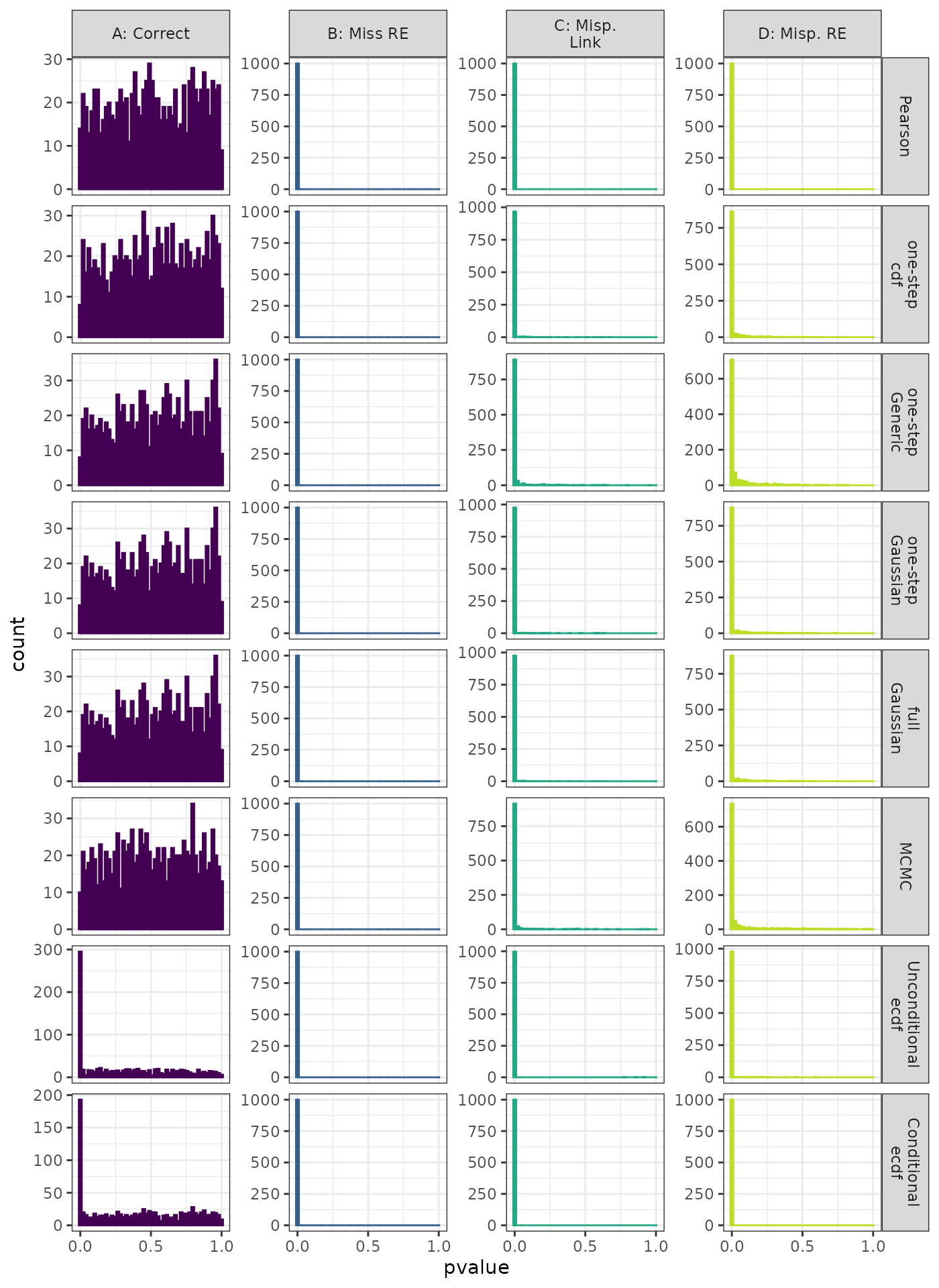
Linear Mixed Model. Distribution of theoretical p-values under the correct (A) and mis-specified (B-D) models evaluated under the Anderson-Darling Test for each method when true parameters are known. Mis-specification are, from left to right: Missing Random Effect (B),, Log-linked data fit to Normal (C), Mis-specified Random Effect (D).
Phylogenetic Autocorrelation Test
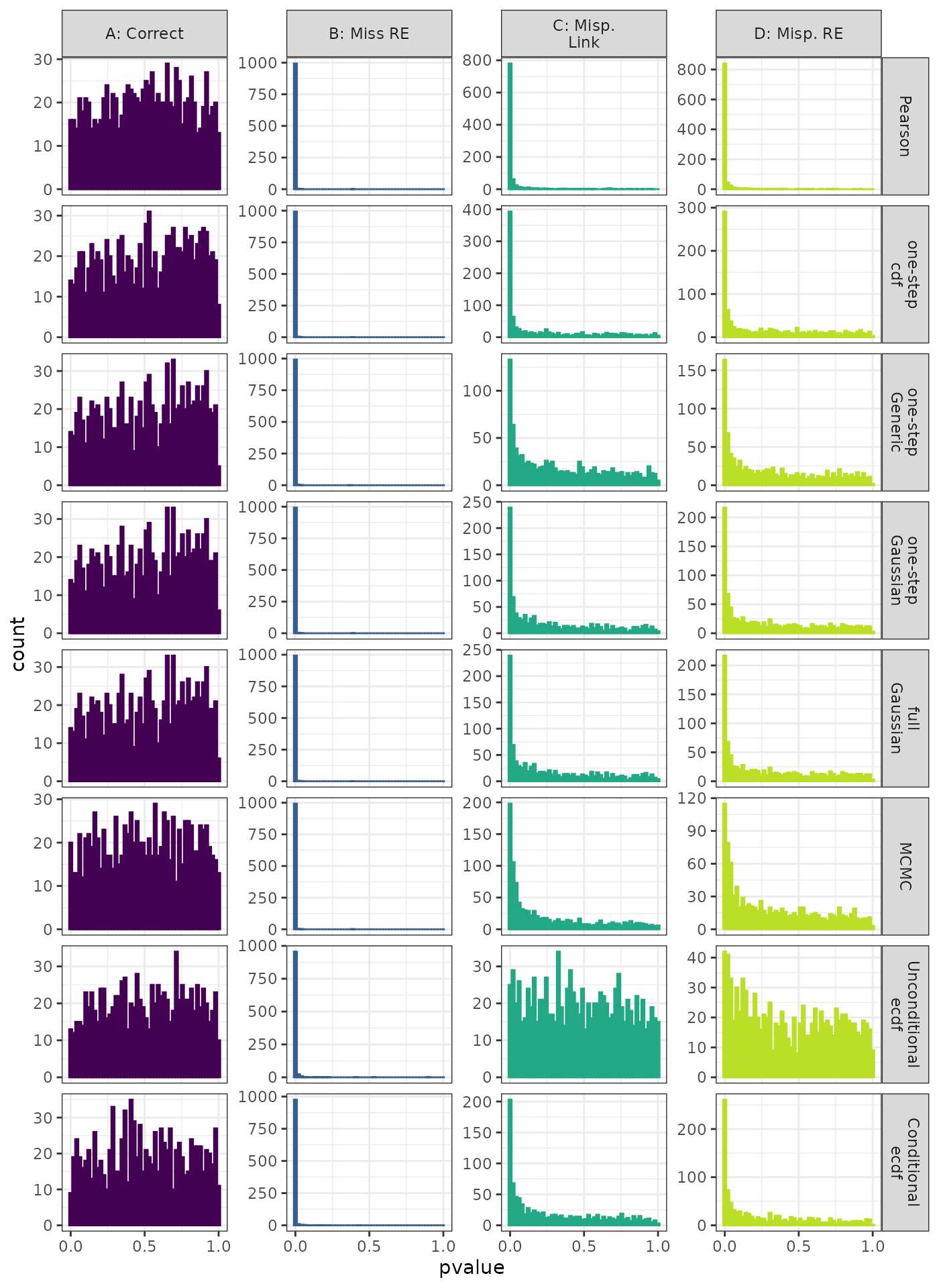
Linear Mixed Model. Distribution of theoretical p-values under the correct (A) and mis-specified (B-D) models evaluated under the Moran’s I Phylogenetic Correlation Test for each method when true parameters are known. Mis-specification are, from left to right: Missing Random Effect (B), Log-linked data fit to Normal (C), Mis-specified Random Effect (D).
Estimated Residuals
Kolmogorov-Smirnov
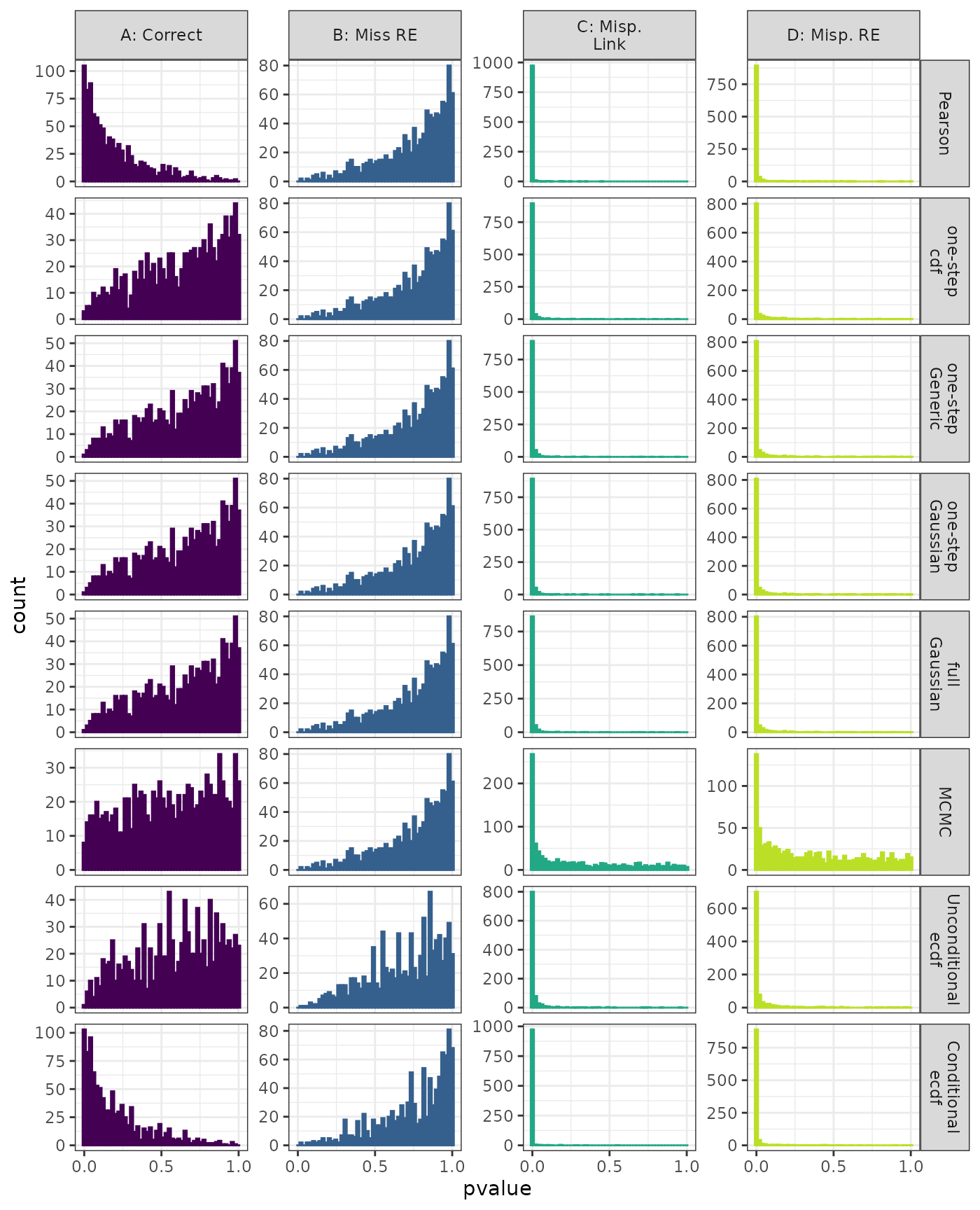
Linear Mixed Model. Distribution of estimated p-values under the correct (A) and mis-specified (B-D) models evaluated under the Kolmogorov-Smirnov Test for each method when parameters are estimated. Mis-specification are, from left to right: Missing Random Effect (B), Log-linked data fit to Normal (C), Mis-specified Random Effect (D).
Lilliefors
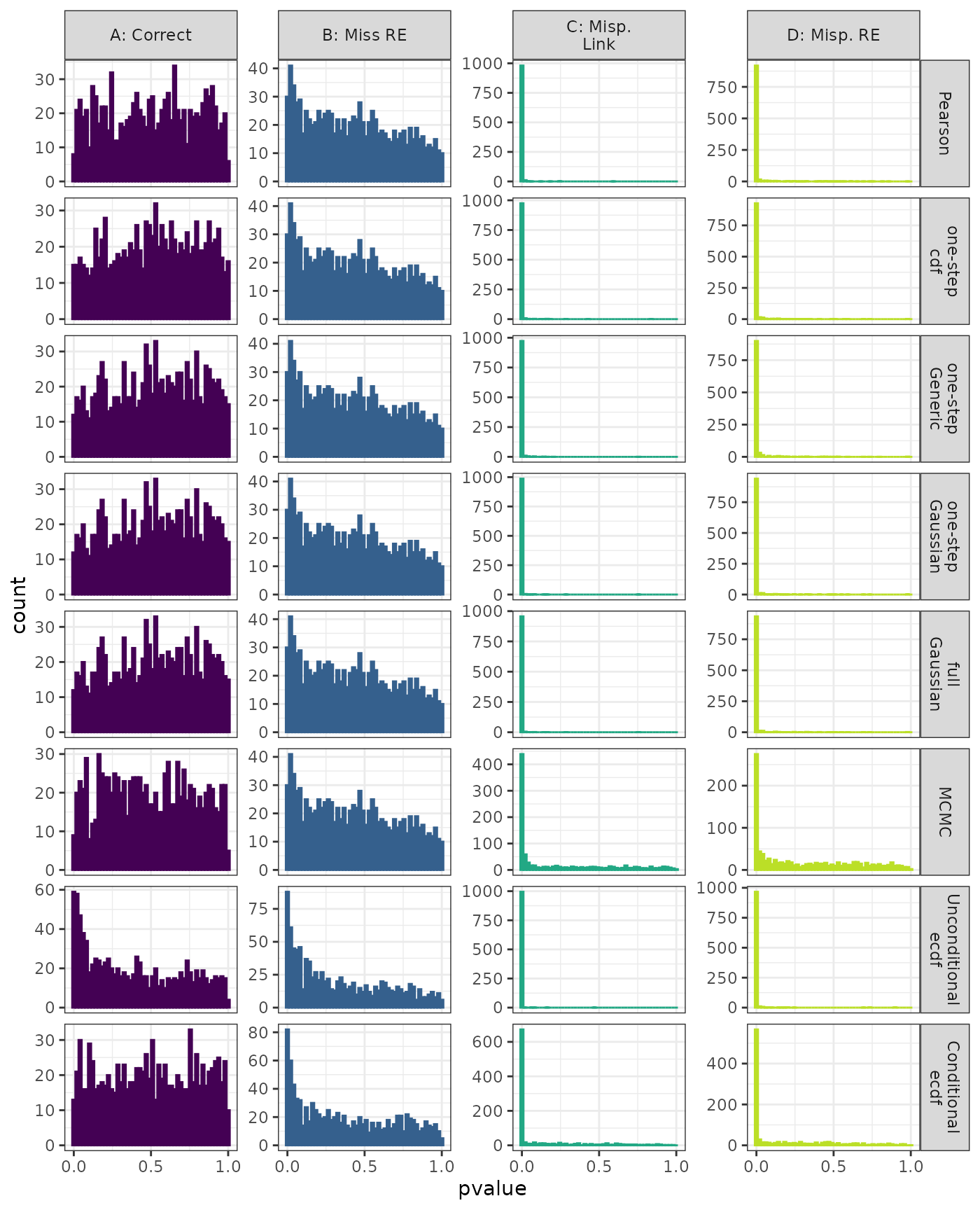
Linear Mixed Model. Distribution of estimated p-values under the correct (A) and mis-specified (B-D) models evaluated under the Lilliefors Test for each method when parameters are estimated. Mis-specification are, from left to right: Missing Random Effect (B), Log-linked data fit to Normal (C), Mis-specified Random Effect (D).
Anderson-Darling Test
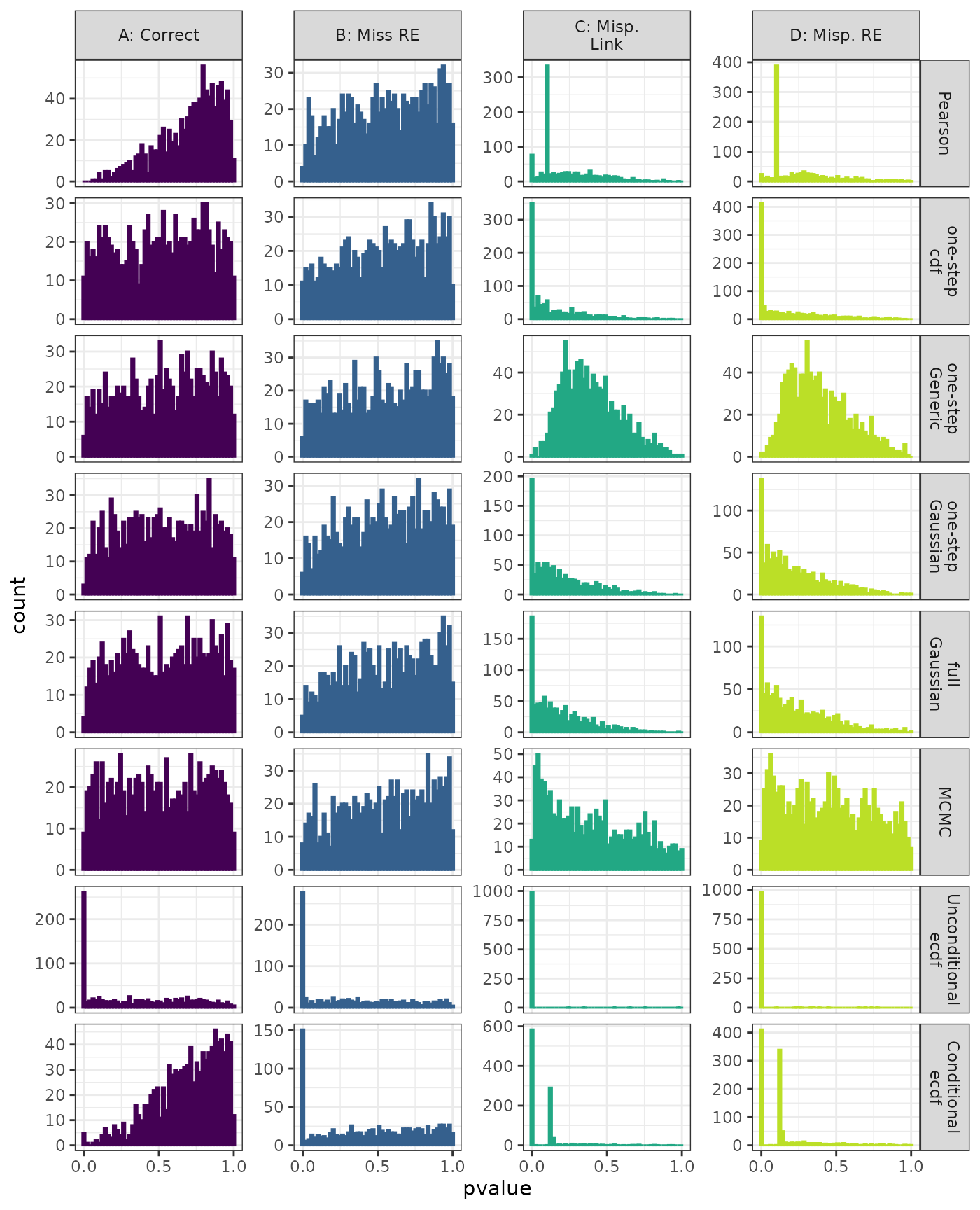
Linear Mixed Model. Distribution of estimated p-values under the correct (A) and mis-specified (B-D) models evaluated under the Anderson-Darling Test for each method when parameters are estimated. Mis-specification are, from left to right: Missing Random Effect (B), Log-linked data fit to Normal (C),Mis-specified Random Effect (D).
Phylogenetic Autocorrelation Test
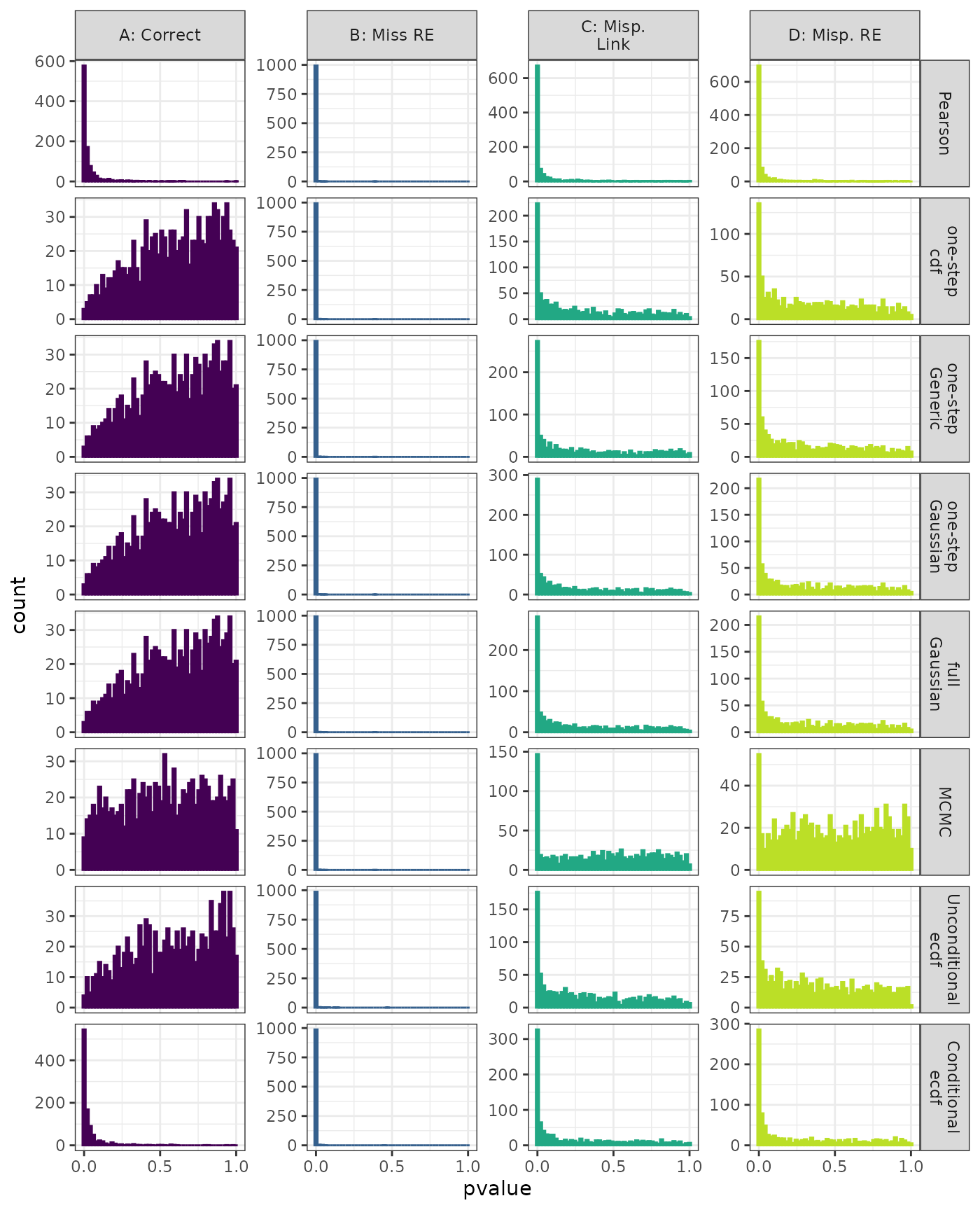
Linear Mixed Model. Distribution of estimated p-values under the correct (A) and mis-specified (B-D) models evaluated under the Moran’s I phylogenetic autocorrelation test for each method when parameters are estimated. Mis-specification are, from left to right: Missing Random Effect (B), Log-linked data fit to Normal (C), Mis-specified Random Effect (D).
Generalized Linear Mixed Model
Theoretical Residuals
Kolmogorov-Smirnov Test
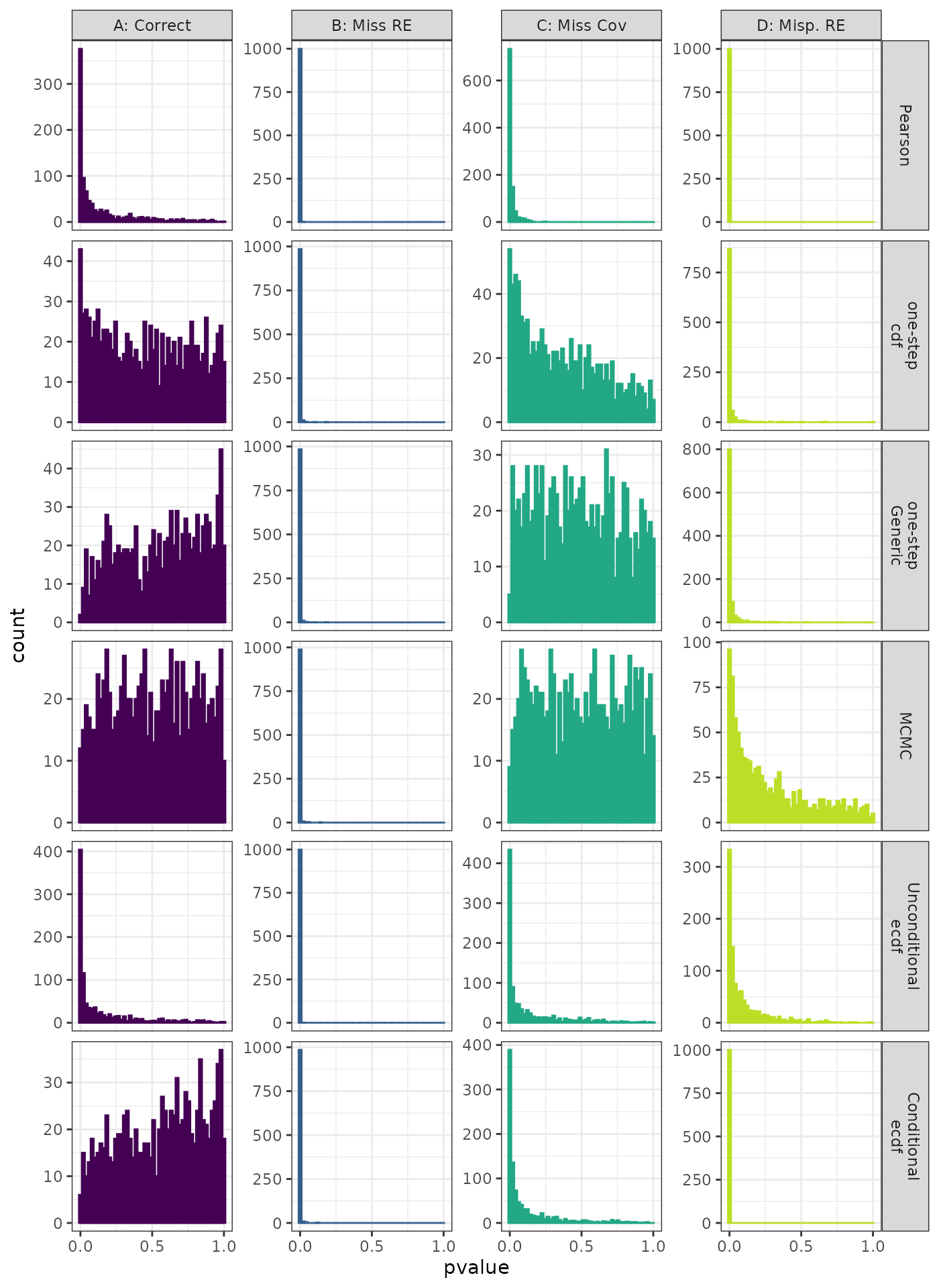
Generalized Linear Mixed Model. Distribution of theoretical p-values under the correct (A) and mis-specified (B-D) models evaluated under the Kolmogorov-Smirnov Test for each method when true parameters are known. Mis-specification are, from left to right: Missing Random Effect (B), Missing Covariate (C), Mis-specified Random Effect (D).
Anderson-Darling Test
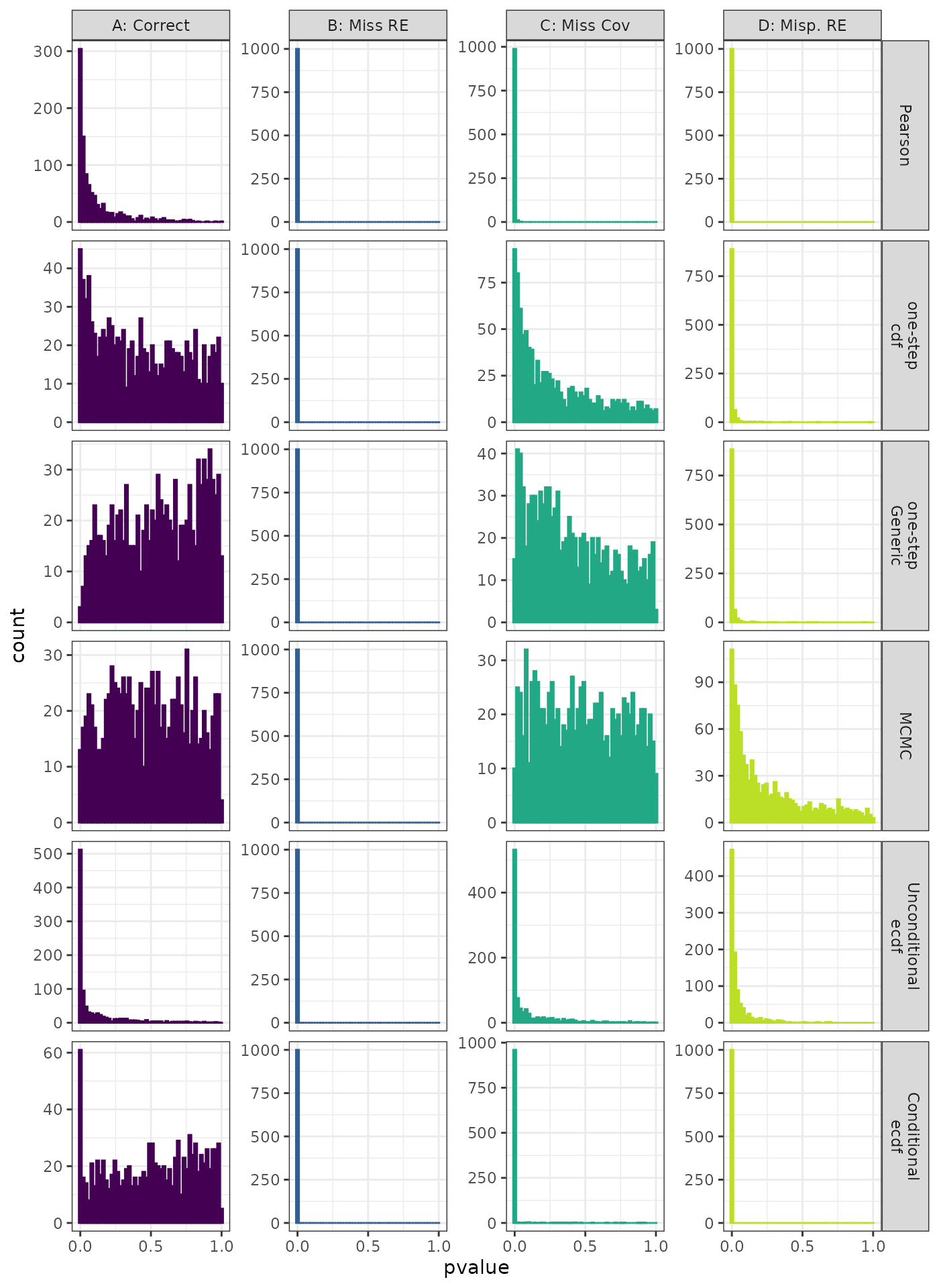
Generalized Linear Mixed Model. Distribution of theoretical p-values under the correct (A) and mis-specified (B-D) models evaluated under the Anderson-Darling Test for each method when true parameters are known. Mis-specification are, from left to right: Missing Random Effect (B), Missing Covariate (C), Mis-specified Random Effect (D).
Phylogenetic Autocorrelation Test
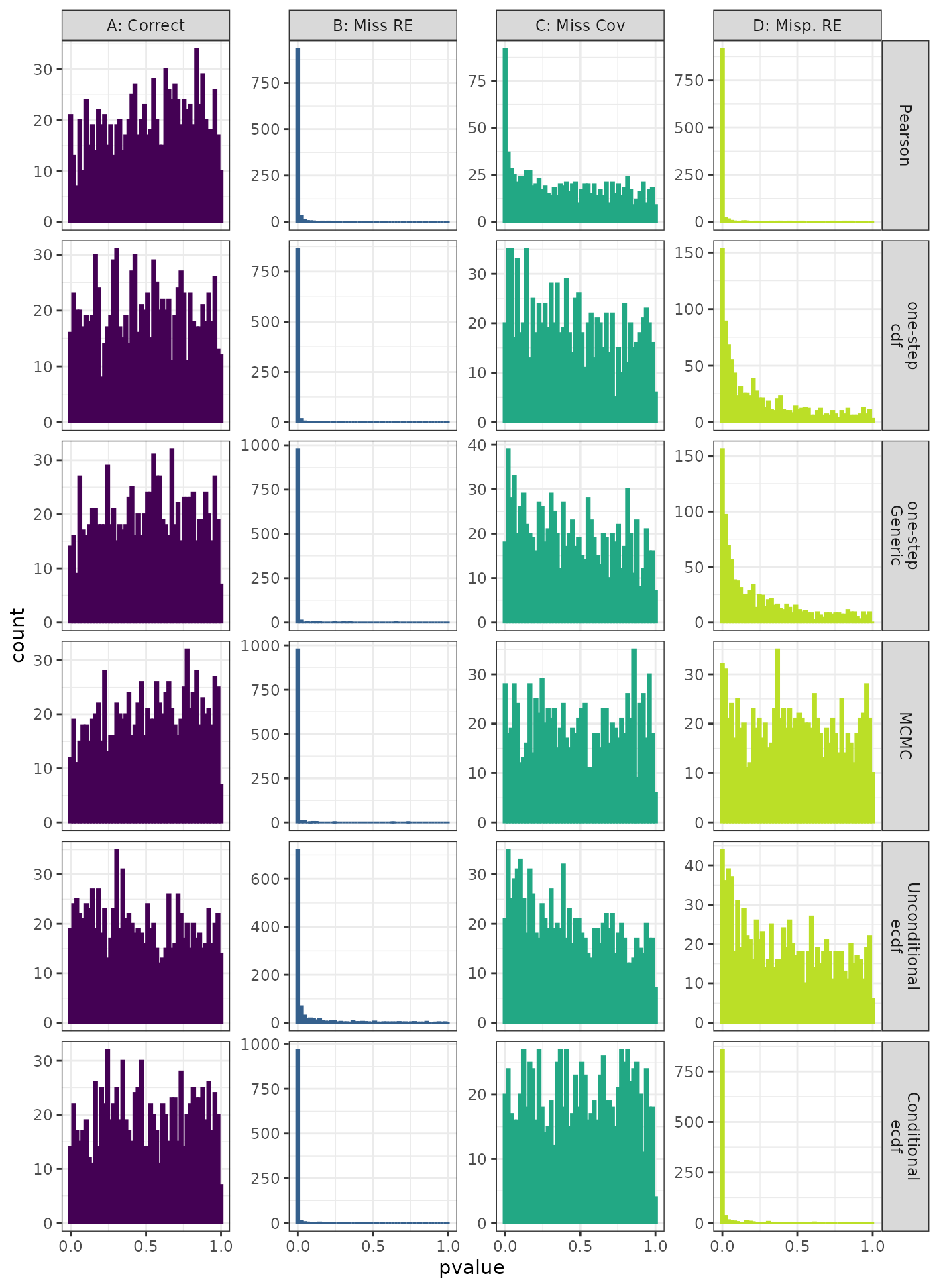
Generalized Linear Mixed Model. Distribution of theoretical p-values under the correct (A) and mis-specified (B-D) models evaluated under the Moran’s I Phylogenetic Correlation Test for each method when true parameters are known. Mis-specification are, from left to right: Missing Random Effect (B), Missing Covariate (C), Mis-specified Random Effect (D).
Estimated Residuals
Kolmogorov-Smirnov
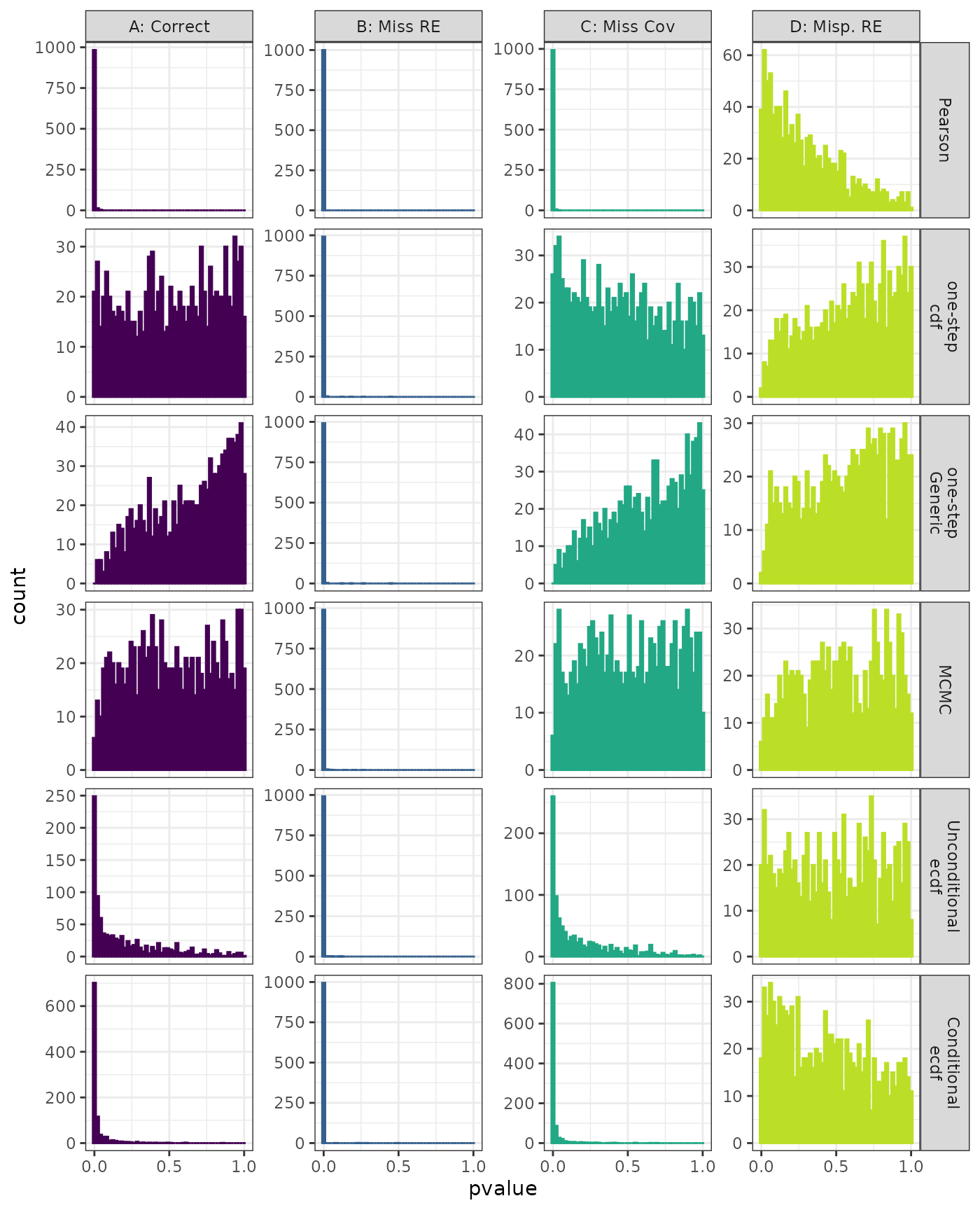
Generalized Linear Mixed Model. Distribution of estimated p-values under the correct (A) and mis-specified (B-D) models evaluated under the Kolmogorov-Smirnov Test for each method when parameters are estimated. Mis-specification are, from left to right: Missing Random Effect (B), Missing Covariate (C), Mis-specified Random Effect (D).
Lilliefors
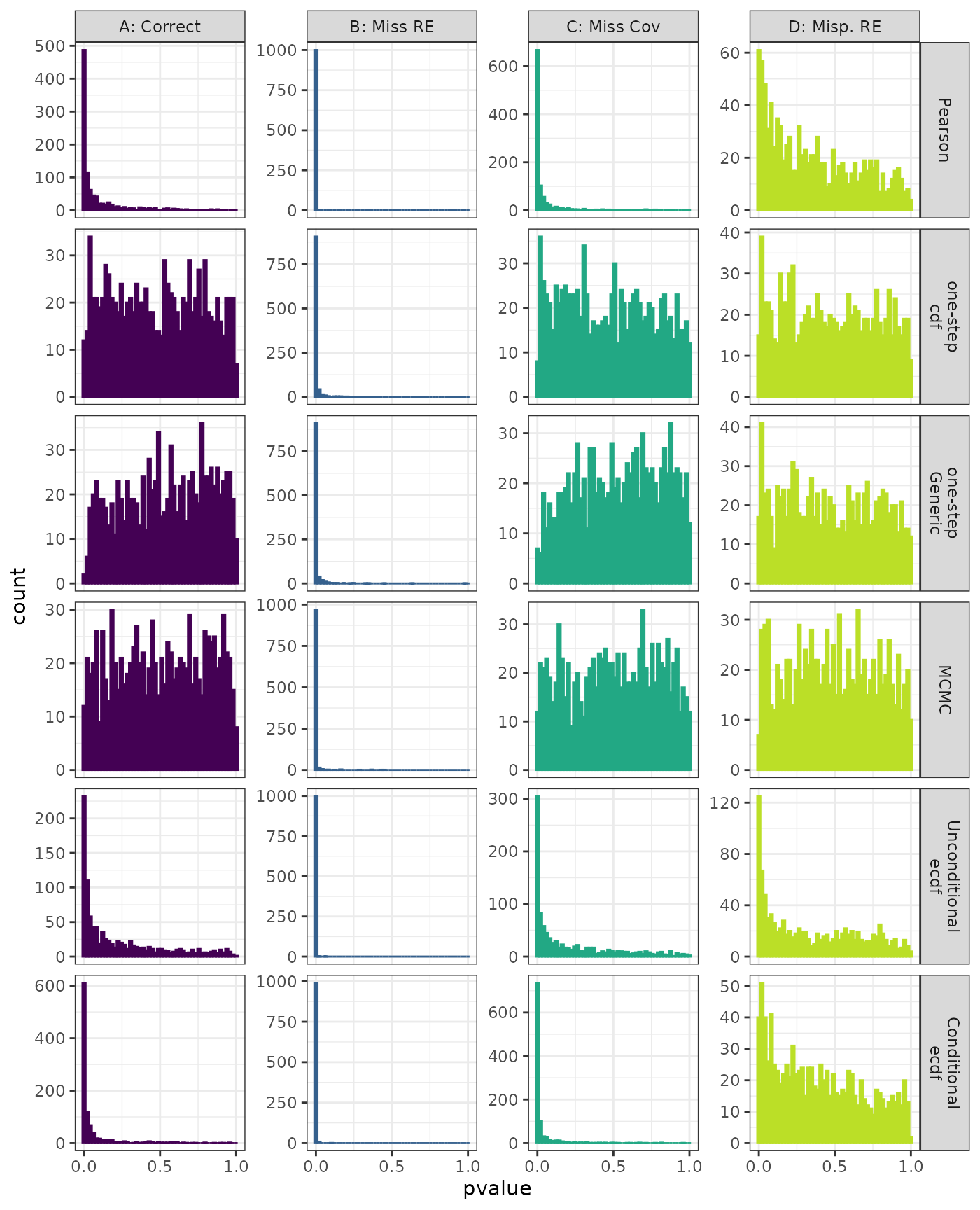
Generalized Linear Mixed Model. Distribution of estimated p-values under the correct (A) and mis-specified (B-D) models evaluated under the Lilliefors Test for each method when parameters are estimated. Mis-specification are, from left to right: Missing Random Effect (B), Missing Covariate (C), Mis-specified Random Effect (D).
Anderson-Darling Test
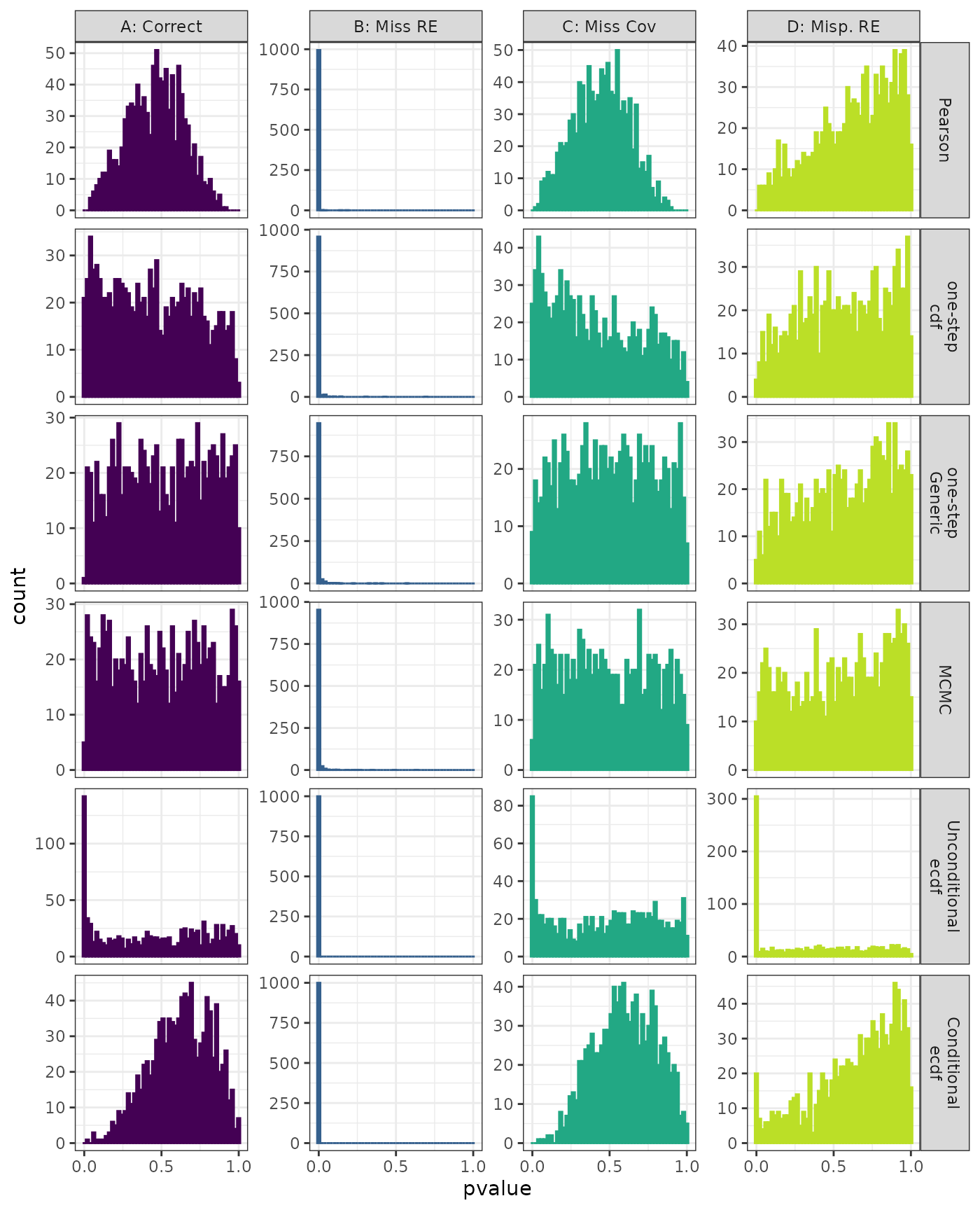
Generalized Linear Mixed Model. Distribution of estimated p-values under the correct (A) and mis-specified (B-D) models evaluated under the Anderson-Darling Test for each method when parameters are estimated. Mis-specification are, from left to right: Missing Random Effect (B), Missing Covariate (C), Mis-specified Random Effect (D).
Phylogenetic Autocorrelation Test
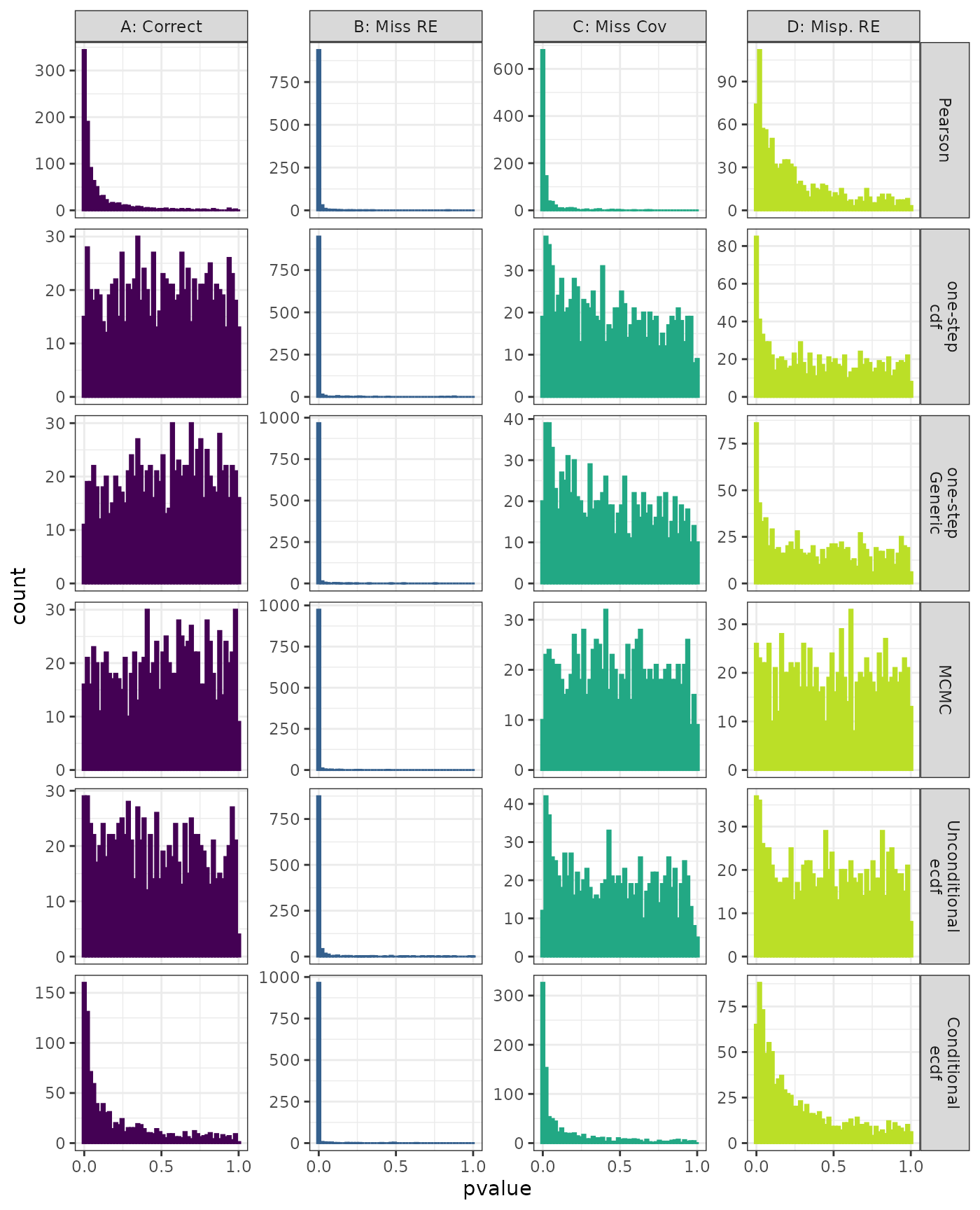
Generalized Linear Mixed Model. Distribution of estimated p-values under the correct (A) and mis-specified (B-D) models evaluated under the Moran’s I phylogenetic autocorrelation test for each method when parameters are estimated. Mis-specification are, from left to right: Missing Random Effect (B), Missing Covariate (C), Mis-specified Random Effect (D).